

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Ligand | BDBM50399766 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_877370 (CHEMBL2182415) | ||
| Ki | 4.7±n/a nM | ||
| Citation |  Mazurov, AA; Kombo, DC; Hauser, TA; Miao, L; Dull, G; Genus, JF; Fedorov, NB; Benson, L; Sidach, S; Xiao, Y; Hammond, PS; James, JW; Miller, CH; Yohannes, D Discovery of (2S,3R)-N-[2-(Pyridin-3-ylmethyl)-1-azabicyclo[2.2.2]oct-3-yl]benzo[b]furan-2-carboxamide (TC-5619), a selectivea7 nicotinic acetylcholine receptor agonist, for the treatment of cognitive disorders. J Med Chem55:9793-809 (2012) [PubMed] Article Mazurov, AA; Kombo, DC; Hauser, TA; Miao, L; Dull, G; Genus, JF; Fedorov, NB; Benson, L; Sidach, S; Xiao, Y; Hammond, PS; James, JW; Miller, CH; Yohannes, D Discovery of (2S,3R)-N-[2-(Pyridin-3-ylmethyl)-1-azabicyclo[2.2.2]oct-3-yl]benzo[b]furan-2-carboxamide (TC-5619), a selectivea7 nicotinic acetylcholine receptor agonist, for the treatment of cognitive disorders. J Med Chem55:9793-809 (2012) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuronal acetylcholine receptor subunit alpha-7 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Synonyms: | ACHA7_HUMAN | CHRNA7 | Cholinergic, Nicotinic Alpha7 | NACHRA7 | Neuronal acetylcholine receptor protein alpha-7 subunit | Neuronal acetylcholine receptor subunit alpha-7 (nAChR-alpha 7) | Nicotinic acetylcholine receptor alpha-7 (alpha7 nAChR) | ||
| Type: | n/a | ||
| Mol. Mass.: | 56448.33 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | CHRNA7 (NACHRA7) | ||
| Residue: | 502 | ||
| Sequence: |
| ||
| BDBM50399766 | |||
| n/a | |||
| Name | BDBM50399766 | ||
| Synonyms: | CHEMBL2180254 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H22FN3O2 | ||
| Mol. Mass. | 379.4274 | ||
| SMILES | Fc1ccc2oc(cc2c1)C(=O)N[C@@H]1C2CCN(CC2)[C@H]1Cc1cccnc1 |r,wU:13.14,wD:20.24,(11.82,-24.93,;11.19,-26.33,;12.09,-27.58,;11.45,-28.98,;9.93,-29.13,;9.02,-30.37,;7.56,-29.88,;7.57,-28.35,;9.04,-27.88,;9.67,-26.49,;6.41,-30.92,;6.41,-32.46,;5.08,-30.14,;3.74,-30.91,;2.41,-30.13,;1.08,-30.91,;1.08,-32.45,;2.41,-33.21,;1.62,-31.88,;3,-31.38,;3.74,-32.45,;5.07,-33.22,;5.07,-34.77,;3.73,-35.53,;3.73,-37.07,;5.07,-37.85,;6.41,-37.07,;6.4,-35.53,)| | ||
| Structure | 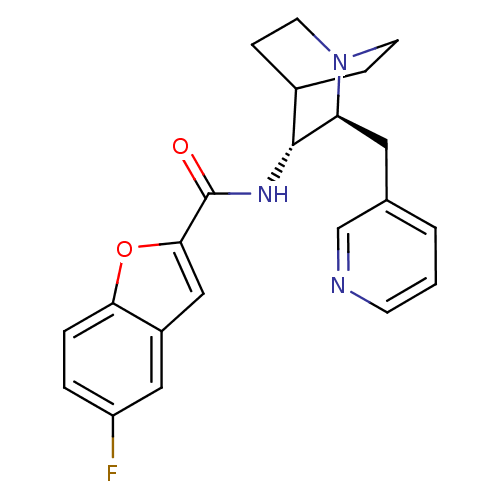
| ||