| Reaction Details |
|---|
 | Report a problem with these data |
| Target | P2X purinoceptor 7 |
|---|
| Ligand | BDBM50410955 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_469376 (CHEMBL932853) |
|---|
| IC50 | 309.03±n/a nM |
|---|
| Citation |  Florjancic, AS; Peddi, S; Perez-Medrano, A; Li, B; Namovic, MT; Grayson, G; Donnelly-Roberts, DL; Jarvis, MF; Carroll, WA Synthesis and in vitro activity of 1-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-1H-1,2,4-triazol-5-amine and 4-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-4H-1,2,4-triazol-3-amine P2X7 antagonists. Bioorg Med Chem Lett18:2089-92 (2008) [PubMed] Article Florjancic, AS; Peddi, S; Perez-Medrano, A; Li, B; Namovic, MT; Grayson, G; Donnelly-Roberts, DL; Jarvis, MF; Carroll, WA Synthesis and in vitro activity of 1-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-1H-1,2,4-triazol-5-amine and 4-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-4H-1,2,4-triazol-3-amine P2X7 antagonists. Bioorg Med Chem Lett18:2089-92 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| P2X purinoceptor 7 |
|---|
| Name: | P2X purinoceptor 7 |
|---|
| Synonyms: | ATP receptor | P2RX7_RAT | P2X purinoceptor 7 | P2X purinoceptor 7 (P2RX7) | P2X purinoceptor 7 (P2X7) | P2X7 | P2X7 rat | P2Z receptor | P2rx7 | Purinergic receptor |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 68403.50 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | Q64663 |
|---|
| Residue: | 595 |
|---|
| Sequence: | MPACCSWNDVFQYETNKVTRIQSVNYGTIKWILHMTVFSYVSFALMSDKLYQRKEPLISS
VHTKVKGVAEVTENVTEGGVTKLVHGIFDTADYTLPLQGNSFFVMTNYLKSEGQEQKLCP
EYPSRGKQCHSDQGCIKGWMDPQSKGIQTGRCIPYDQKRKTCEIFAWCPAEEGKEAPRPA
LLRSAENFTVLIKNNIDFPGHNYTTRNILPGMNISCTFHKTWNPQCPIFRLGDIFQEIGE
NFTEVAVQGGIMGIEIYWDCNLDSWSHRCQPKYSFRRLDDKYTNESLFPGYNFRYAKYYK
ENGMEKRTLIKAFGVRFDILVFGTGGKFDIIQLVVYIGSTLSYFGLATVCIDLIINTYAS
TCCRSRVYPSCKCCEPCAVNEYYYRKKCEPIVEPKPTLKYVSFVDEPHIWMVDQQLLGKS
LQDVKGQEVPRPQTDFLELSRLSLSLHHSPPIPGQPEEMQLLQIEAVPRSRDSPDWCQCG
NCLPSQLPENRRALEELCCRRKPGQCITTSELFSKIVLSREALQLLLLYQEPLLALEGEA
INSKLRHCAYRSYATWRFVSQDMADFAILPSCCRWKIRKEFPKTQGQYSGFKYPY
|
|
|
|---|
| BDBM50410955 |
|---|
| n/a |
|---|
| Name | BDBM50410955 |
|---|
| Synonyms: | A-438079 | CHEMBL377219 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H9Cl2N5 |
|---|
| Mol. Mass. | 306.15 |
|---|
| SMILES | Clc1cccc(-c2nnnn2Cc2cccnc2)c1Cl |
|---|
| Structure | 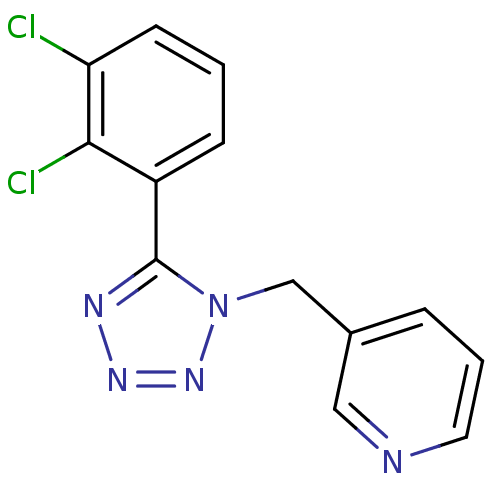
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Florjancic, AS; Peddi, S; Perez-Medrano, A; Li, B; Namovic, MT; Grayson, G; Donnelly-Roberts, DL; Jarvis, MF; Carroll, WA Synthesis and in vitro activity of 1-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-1H-1,2,4-triazol-5-amine and 4-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-4H-1,2,4-triazol-3-amine P2X7 antagonists. Bioorg Med Chem Lett18:2089-92 (2008) [PubMed] Article
Florjancic, AS; Peddi, S; Perez-Medrano, A; Li, B; Namovic, MT; Grayson, G; Donnelly-Roberts, DL; Jarvis, MF; Carroll, WA Synthesis and in vitro activity of 1-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-1H-1,2,4-triazol-5-amine and 4-(2,3-dichlorophenyl)-N-(pyridin-3-ylmethyl)-4H-1,2,4-triazol-3-amine P2X7 antagonists. Bioorg Med Chem Lett18:2089-92 (2008) [PubMed] Article