| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM50132713 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_592876 (CHEMBL1046621) |
|---|
| IC50 | 0.331±n/a nM |
|---|
| Citation |  Kapanda, CN; Muccioli, GG; Labar, G; Poupaert, JH; Lambert, DM Bis(dialkylaminethiocarbonyl)disulfides as potent and selective monoglyceride lipase inhibitors. J Med Chem52:7310-4 (2009) [PubMed] Article Kapanda, CN; Muccioli, GG; Labar, G; Poupaert, JH; Lambert, DM Bis(dialkylaminethiocarbonyl)disulfides as potent and selective monoglyceride lipase inhibitors. J Med Chem52:7310-4 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM50132713 |
|---|
| n/a |
|---|
| Name | BDBM50132713 |
|---|
| Synonyms: | Arachidonic acid derivative | CHEMBL113262 | Methyl arachidonoyl fluorophophonate | methyl ((5Z,8Z,11Z,14Z)-2-Icosa-5,8,11,14-tetraenyl)phosphonofluoridoate | methyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenylphosphonofluoridate | methyl -icosa-5,8,11,14-tetraenylphosphonofluoridate | methyl(5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenylphosphonofluoridate | methylarachidonyl fluorophosphonate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H36FO2P |
|---|
| Mol. Mass. | 370.4815 |
|---|
| SMILES | CCCCC\C=C/C\C=C/C\C=C/C\C=C/CCCCP(F)(=O)OC |
|---|
| Structure | 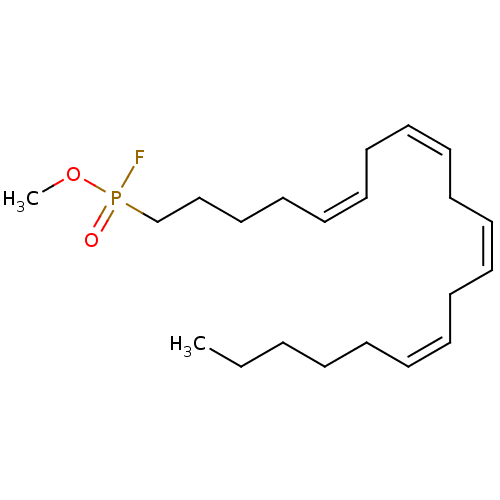
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kapanda, CN; Muccioli, GG; Labar, G; Poupaert, JH; Lambert, DM Bis(dialkylaminethiocarbonyl)disulfides as potent and selective monoglyceride lipase inhibitors. J Med Chem52:7310-4 (2009) [PubMed] Article
Kapanda, CN; Muccioli, GG; Labar, G; Poupaert, JH; Lambert, DM Bis(dialkylaminethiocarbonyl)disulfides as potent and selective monoglyceride lipase inhibitors. J Med Chem52:7310-4 (2009) [PubMed] Article