| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase sirtuin-1 |
|---|
| Ligand | BDBM50425824 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_937895 (CHEMBL2317035) |
|---|
| Kd | 1300±n/a nM |
|---|
| Citation |  Zhao, X; Allison, D; Condon, B; Zhang, F; Gheyi, T; Zhang, A; Ashok, S; Russell, M; MacEwan, I; Qian, Y; Jamison, JA; Luz, JG The 2.5Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem56:963-9 (2013) [PubMed] Article Zhao, X; Allison, D; Condon, B; Zhang, F; Gheyi, T; Zhang, A; Ashok, S; Russell, M; MacEwan, I; Qian, Y; Jamison, JA; Luz, JG The 2.5Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem56:963-9 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase sirtuin-1 |
|---|
| Name: | NAD-dependent protein deacetylase sirtuin-1 |
|---|
| Synonyms: | NAD-Dependent Deacetylase Sirtuin-1 | NAD-dependent deacetylase sirtuin 1 | NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | SIR1_HUMAN | SIR2-like protein 1 | SIR2L1 | SIRT1 | Sirtuin 1 (SIRT1) | Sirtuin-1 (SIRT1) | hSIR2 |
|---|
| Type: | Developmental protein; hydrolase |
|---|
| Mol. Mass.: | 81626.66 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 747 |
|---|
| Sequence: | MADEAALALQPGGSPSAAGADREAASSPAGEPLRKRPRRDGPGLERSPGEPGGAAPEREV
PAAARGCPGAAAAALWREAEAEAAAAGGEQEAQATAAAGEGDNGPGLQGPSREPPLADNL
YDEDDDDEGEEEEEAAAAAIGYRDNLLFGDEIITNGFHSCESDEEDRASHASSSDWTPRP
RIGPYTFVQQHLMIGTDPRTILKDLLPETIPPPELDDMTLWQIVINILSEPPKRKKRKDI
NTIEDAVKLLQECKKIIVLTGAGVSVSCGIPDFRSRDGIYARLAVDFPDLPDPQAMFDIE
YFRKDPRPFFKFAKEIYPGQFQPSLCHKFIALSDKEGKLLRNYTQNIDTLEQVAGIQRII
QCHGSFATASCLICKYKVDCEAVRGDIFNQVVPRCPRCPADEPLAIMKPEIVFFGENLPE
QFHRAMKYDKDEVDLLIVIGSSLKVRPVALIPSSIPHEVPQILINREPLPHLHFDVELLG
DCDVIINELCHRLGGEYAKLCCNPVKLSEITEKPPRTQKELAYLSELPPTPLHVSEDSSS
PERTSPPDSSVIVTLLDQAAKSNDDLDVSESKGCMEEKPQEVQTSRNVESIAEQMENPDL
KNVGSSTGEKNERTSVAGTVRKCWPNRVAKEQISRRLDGNQYLFLPPNRYIFHGAEVYSD
SEDDVLSSSSCGSNSDSGTCQSPSLEEPMEDESEIEEFYNGLEDEPDVPERAGGAGFGTD
GDDQEAINEAISVKQEVTDMNYPSNKS
|
|
|
|---|
| BDBM50425824 |
|---|
| n/a |
|---|
| Name | BDBM50425824 |
|---|
| Synonyms: | CHEMBL2312289 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H13ClN2O |
|---|
| Mol. Mass. | 248.708 |
|---|
| SMILES | NC(=O)[C@@H]1CCCc2c1[nH]c1ccc(Cl)cc21 |r| |
|---|
| Structure | 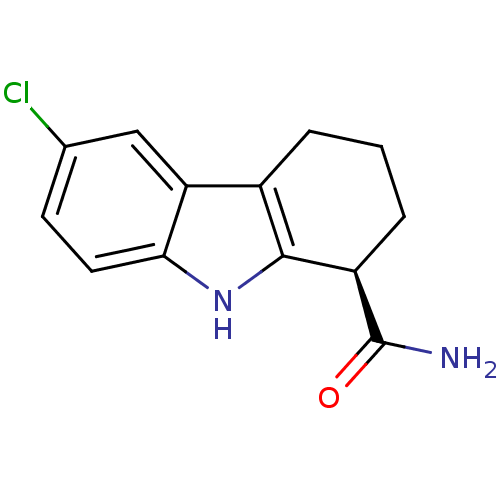
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Zhao, X; Allison, D; Condon, B; Zhang, F; Gheyi, T; Zhang, A; Ashok, S; Russell, M; MacEwan, I; Qian, Y; Jamison, JA; Luz, JG The 2.5Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem56:963-9 (2013) [PubMed] Article
Zhao, X; Allison, D; Condon, B; Zhang, F; Gheyi, T; Zhang, A; Ashok, S; Russell, M; MacEwan, I; Qian, Y; Jamison, JA; Luz, JG The 2.5Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem56:963-9 (2013) [PubMed] Article