

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Phospholipase A2, membrane associated | ||
| Ligand | BDBM50428491 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_943475 (CHEMBL2342415) | ||
| IC50 | 155000±n/a nM | ||
| Citation |  Ye, L; Dickerson, T; Kaur, H; Takada, YK; Fujita, M; Liu, R; Knapp, JM; Lam, KS; Schore, NE; Kurth, MJ; Takada, Y Identification of inhibitors against interaction between pro-inflammatory sPLA2-IIA protein and integrinavß3. Bioorg Med Chem Lett23:340-5 (2012) [PubMed] Article Ye, L; Dickerson, T; Kaur, H; Takada, YK; Fujita, M; Liu, R; Knapp, JM; Lam, KS; Schore, NE; Kurth, MJ; Takada, Y Identification of inhibitors against interaction between pro-inflammatory sPLA2-IIA protein and integrinavß3. Bioorg Med Chem Lett23:340-5 (2012) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Phospholipase A2, membrane associated | |||
| Name: | Phospholipase A2, membrane associated | ||
| Synonyms: | GIIC sPLA2 | Group IIA phospholipase A2 | NPS-PLA2 | Non-Pancreatic Secretory Phospholipase A2 | Non-pancreatic secretory phospholipase A2 (hnps-PLA2) | PA2GA_HUMAN | PLA2B | PLA2G2A | PLA2L | Phosphatidylcholine 2-acylhydrolase | Phospholipase A2 group IIA | RASF-A | ||
| Type: | Hydrolase | ||
| Mol. Mass.: | 16101.20 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | The human nps PLA2 was cloned, and expressed in E. coli. There was a refolding process in the purification. | ||
| Residue: | 144 | ||
| Sequence: |
| ||
| BDBM50428491 | |||
| n/a | |||
| Name | BDBM50428491 | ||
| Synonyms: | CHEMBL2335198 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H35N11O9S | ||
| Mol. Mass. | 713.721 | ||
| SMILES | Cc1nc(NC(=O)c2ccccc2)sc1-c1cc([nH]n1)C(=O)NCC(=O)N[C@H](CCCN=C(N)N)C(=O)NCC(=O)N[C@H](CC(O)=O)C(O)=O |r,wU:27.29,42.44,(6.91,-18.61,;5.37,-18.45,;4.35,-19.6,;2.94,-18.97,;1.61,-19.74,;.27,-18.97,;.27,-17.43,;-1.07,-19.74,;-2.4,-18.97,;-3.73,-19.74,;-3.73,-21.28,;-2.38,-22.05,;-1.06,-21.28,;3.1,-17.43,;4.61,-17.11,;5.23,-15.7,;6.73,-15.38,;6.89,-13.85,;5.49,-13.23,;4.46,-14.37,;8.22,-13.08,;8.22,-11.54,;9.56,-13.85,;10.89,-13.07,;12.22,-13.85,;12.22,-15.39,;13.56,-13.07,;14.89,-13.85,;14.89,-15.39,;16.23,-16.15,;16.23,-17.69,;17.57,-18.47,;17.57,-20.01,;16.23,-20.78,;18.9,-20.78,;16.23,-13.07,;16.23,-11.53,;17.57,-13.85,;18.9,-13.07,;20.24,-13.85,;20.24,-15.39,;21.57,-13.07,;22.91,-13.85,;22.91,-15.39,;24.24,-16.15,;25.57,-15.39,;24.24,-17.69,;24.24,-13.07,;25.56,-13.84,;24.24,-11.53,)| | ||
| Structure | 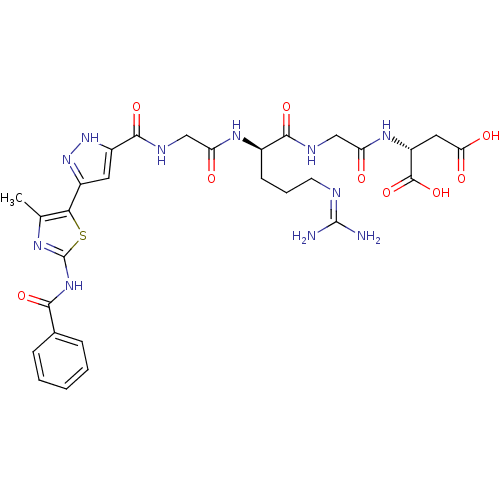
| ||