| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mineralocorticoid receptor |
|---|
| Ligand | BDBM50448165 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1292841 (CHEMBL3122505) |
|---|
| Ki | 2.5±n/a nM |
|---|
| Citation |  Carson, MW; Luz, JG; Suen, C; Montrose, C; Zink, R; Ruan, X; Cheng, C; Cole, H; Adrian, MD; Kohlman, DT; Mabry, T; Snyder, N; Condon, B; Maletic, M; Clawson, D; Pustilnik, A; Coghlan, MJ Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design. J Med Chem57:849-60 (2014) [PubMed] Article Carson, MW; Luz, JG; Suen, C; Montrose, C; Zink, R; Ruan, X; Cheng, C; Cole, H; Adrian, MD; Kohlman, DT; Mabry, T; Snyder, N; Condon, B; Maletic, M; Clawson, D; Pustilnik, A; Coghlan, MJ Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design. J Med Chem57:849-60 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mineralocorticoid receptor |
|---|
| Name: | Mineralocorticoid receptor |
|---|
| Synonyms: | MCR | MCR_HUMAN | MLR | MR | NR3C2 | Nuclear receptor subfamily 3 group C member 2 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 107076.42 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08235 |
|---|
| Residue: | 984 |
|---|
| Sequence: | METKGYHSLPEGLDMERRWGQVSQAVERSSLGPTERTDENNYMEIVNVSCVSGAIPNNST
QGSSKEKQELLPCLQQDNNRPGILTSDIKTELESKELSATVAESMGLYMDSVRDADYSYE
QQNQQGSMSPAKIYQNVEQLVKFYKGNGHRPSTLSCVNTPLRSFMSDSGSSVNGGVMRAV
VKSPIMCHEKSPSVCSPLNMTSSVCSPAGINSVSSTTASFGSFPVHSPITQGTPLTCSPN
VENRGSRSHSPAHASNVGSPLSSPLSSMKSSISSPPSHCSVKSPVSSPNNVTLRSSVSSP
ANINNSRCSVSSPSNTNNRSTLSSPAASTVGSICSPVNNAFSYTASGTSAGSSTLRDVVP
SPDTQEKGAQEVPFPKTEEVESAISNGVTGQLNIVQYIKPEPDGAFSSSCLGGNSKINSD
SSFSVPIKQESTKHSCSGTSFKGNPTVNPFPFMDGSYFSFMDDKDYYSLSGILGPPVPGF
DGNCEGSGFPVGIKQEPDDGSYYPEASIPSSAIVGVNSGGQSFHYRIGAQGTISLSRSAR
DQSFQHLSSFPPVNTLVESWKSHGDLSSRRSDGYPVLEYIPENVSSSTLRSVSTGSSRPS
KICLVCGDEASGCHYGVVTCGSCKVFFKRAVEGQHNYLCAGRNDCIIDKIRRKNCPACRL
QKCLQAGMNLGARKSKKLGKLKGIHEEQPQQQQPPPPPPPPQSPEEGTTYIAPAKEPSVN
TALVPQLSTISRALTPSPVMVLENIEPEIVYAGYDSSKPDTAENLLSTLNRLAGKQMIQV
VKWAKVLPGFKNLPLEDQITLIQYSWMCLSSFALSWRSYKHTNSQFLYFAPDLVFNEEKM
HQSAMYELCQGMHQISLQFVRLQLTFEEYTIMKVLLLLSTIPKDGLKSQAAFEEMRTNYI
KELRKMVTKCPNNSGQSWQRFYQLTKLLDSMHDLVSDLLEFCFYTFRESHALKVEFPAML
VEIISDQLPKVESGNAKPLYFHRK
|
|
|
|---|
| BDBM50448165 |
|---|
| n/a |
|---|
| Name | BDBM50448165 |
|---|
| Synonyms: | CHEMBL3120320 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H21NO2S |
|---|
| Mol. Mass. | 375.483 |
|---|
| SMILES | [#6]S(=O)(=O)[#7]-c1cccc(\[#6]=[#6]-2/c3ccccc3-[#6]-[#6]-c3ccccc-23)c1 |
|---|
| Structure | 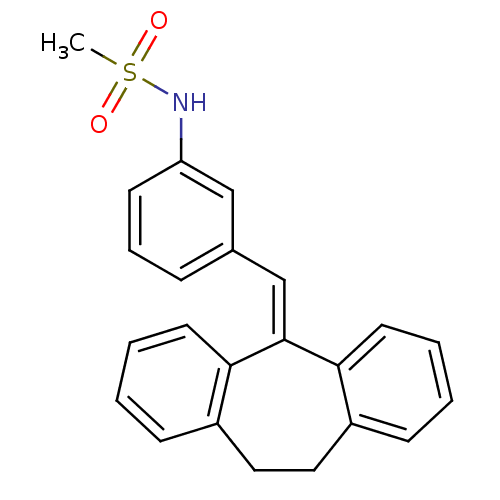
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Carson, MW; Luz, JG; Suen, C; Montrose, C; Zink, R; Ruan, X; Cheng, C; Cole, H; Adrian, MD; Kohlman, DT; Mabry, T; Snyder, N; Condon, B; Maletic, M; Clawson, D; Pustilnik, A; Coghlan, MJ Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design. J Med Chem57:849-60 (2014) [PubMed] Article
Carson, MW; Luz, JG; Suen, C; Montrose, C; Zink, R; Ruan, X; Cheng, C; Cole, H; Adrian, MD; Kohlman, DT; Mabry, T; Snyder, N; Condon, B; Maletic, M; Clawson, D; Pustilnik, A; Coghlan, MJ Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design. J Med Chem57:849-60 (2014) [PubMed] Article