| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Indolethylamine N-methyltransferase |
|---|
| Ligand | BDBM50000193 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1333667 (CHEMBL3231053) |
|---|
| IC50 | >10300±n/a nM |
|---|
| Citation |  Rokach, J; Hamel, P; Hunter, NR; Reader, G; Rooney, CS; Anderson, PS; Cragoe, EJ; Mandel, LR Cyclic amidine inhibitors of indolamine N-methyltransferase. J Med Chem22:237-47 (1979) [PubMed] Rokach, J; Hamel, P; Hunter, NR; Reader, G; Rooney, CS; Anderson, PS; Cragoe, EJ; Mandel, LR Cyclic amidine inhibitors of indolamine N-methyltransferase. J Med Chem22:237-47 (1979) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Indolethylamine N-methyltransferase |
|---|
| Name: | Indolethylamine N-methyltransferase |
|---|
| Synonyms: | Amine N-methyltransferase | Aromatic alkylamine N-methyltransferase | Arylamine N-methyltransferase | INMT | INMT_RABIT | Indolamine N-methyltransferase | Thioether S-methyltransferase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 28949.17 |
|---|
| Organism: | Oryctolagus cuniculus |
|---|
| Description: | ChEMBL_108264 |
|---|
| Residue: | 263 |
|---|
| Sequence: | MEGGFTGGDEYQKHFLPRDYLNTYYSFQSGPSPEAEMLKFNLECLHKTFGPGGLQGDTLI
DIGSGPTIYQVLAACESFKDITLSDFTDRNREELAKWLKKEPGAYDWTPALKFACELEGN
SGRWQEKAEKLRATVKRVLKCDANLSNPLTPVVLPPADCVLTLLAMECACCSLDAYRAAL
RNLASLLKPGGHLVTTVTLQLSSYMVGEREFSCVALEKEEVEQAVLDAGFDIEQLLYSPQ
SYSASTAPNRGVCFLVARKKPGS
|
|
|
|---|
| BDBM50000193 |
|---|
| n/a |
|---|
| Name | BDBM50000193 |
|---|
| Synonyms: | CHEMBL3228379 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H16N2O4 |
|---|
| Mol. Mass. | 228.245 |
|---|
| SMILES | OC(=O)C(O)=O.CN1CCC\C1=N/C1CC1 |
|---|
| Structure | 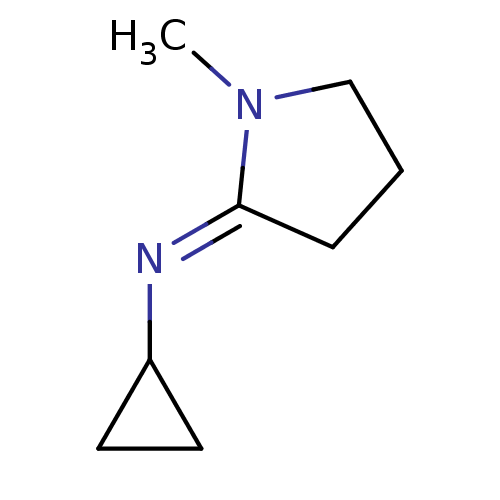
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rokach, J; Hamel, P; Hunter, NR; Reader, G; Rooney, CS; Anderson, PS; Cragoe, EJ; Mandel, LR Cyclic amidine inhibitors of indolamine N-methyltransferase. J Med Chem22:237-47 (1979) [PubMed]
Rokach, J; Hamel, P; Hunter, NR; Reader, G; Rooney, CS; Anderson, PS; Cragoe, EJ; Mandel, LR Cyclic amidine inhibitors of indolamine N-methyltransferase. J Med Chem22:237-47 (1979) [PubMed]