

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neurotensin receptor type 2 | ||
| Ligand | BDBM50019419 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1364514 (CHEMBL3295311) | ||
| EC50 | 68±n/a nM | ||
| Citation |  Thomas, JB; Giddings, AM; Wiethe, RW; Olepu, S; Warner, KR; Sarret, P; Gendron, L; Longpre, JM; Zhang, Y; Runyon, SP; Gilmour, BP Identification of 1-({[1-(4-fluorophenyl)-5-(2-methoxyphenyl)-1H-pyrazol-3-yl]carbonyl}amino)cyclohexane carboxylic acid as a selective nonpeptide neurotensin receptor type 2 compound. J Med Chem57:5318-32 (2014) [PubMed] Article Thomas, JB; Giddings, AM; Wiethe, RW; Olepu, S; Warner, KR; Sarret, P; Gendron, L; Longpre, JM; Zhang, Y; Runyon, SP; Gilmour, BP Identification of 1-({[1-(4-fluorophenyl)-5-(2-methoxyphenyl)-1H-pyrazol-3-yl]carbonyl}amino)cyclohexane carboxylic acid as a selective nonpeptide neurotensin receptor type 2 compound. J Med Chem57:5318-32 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neurotensin receptor type 2 | |||
| Name: | Neurotensin receptor type 2 | ||
| Synonyms: | High-affinity levocabastine-sensitive neurotensin receptor | NT-R-2 | NTR2_RAT | Neurotensin receptor 2 | Neurotensin receptor type 2 | Ntr2 | Ntsr2 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 46280.50 | ||
| Organism: | Rattus norvegicus | ||
| Description: | ChEMBL_1466615 | ||
| Residue: | 416 | ||
| Sequence: |
| ||
| BDBM50019419 | |||
| n/a | |||
| Name | BDBM50019419 | ||
| Synonyms: | CHEMBL3290103 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H30FN3O5 | ||
| Mol. Mass. | 519.564 | ||
| SMILES | COc1cccc(OC)c1-c1cc(nn1-c1ccc(F)cc1)C(=O)NC1(C2CC3CC(C2)CC1C3)C(O)=O |TLB:34:33:31:27.28.29,24:25:27.34.28:32.30.31,THB:34:28:25.33.32:31,29:28:25:32.30.31,29:30:25:27.34.28,35:25:27.34.28:32.30.31,24:25:31:27.28.29,(56.32,-26.53,;57,-27.91,;56.16,-29.2,;54.62,-29.11,;53.77,-30.4,;54.48,-31.78,;56.01,-31.87,;56.7,-33.24,;55.87,-34.52,;56.85,-30.58,;58.38,-30.66,;58.85,-32.13,;60.39,-32.13,;60.87,-30.67,;59.62,-29.75,;59.62,-28.21,;60.96,-27.45,;60.96,-25.91,;59.62,-25.13,;59.62,-23.59,;58.28,-25.92,;58.29,-27.45,;61.3,-33.38,;60.67,-34.79,;62.83,-33.22,;63.74,-34.46,;64.93,-33.19,;66.26,-33.67,;67.66,-33.33,;67.67,-31.8,;66.27,-31.22,;64.92,-31.7,;65.23,-32.45,;65.24,-34.04,;66.65,-34.6,;63.1,-35.87,;61.57,-36.03,;64.01,-37.12,)| | ||
| Structure | 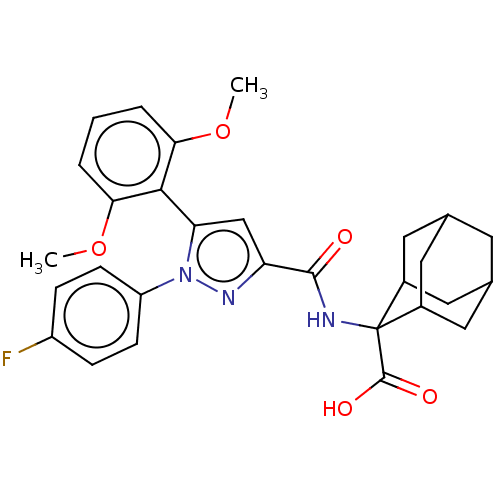
| ||