| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Potassium voltage-gated channel subfamily E/KQT member 1 |
|---|
| Ligand | BDBM50103642 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1478585 (CHEMBL3430486) |
|---|
| IC50 | 1258925±n/a nM |
|---|
| Citation |  Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods70:246-54 (2014) [PubMed] Article Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods70:246-54 (2014) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Potassium voltage-gated channel subfamily E/KQT member 1 |
|---|
| Name: | Potassium voltage-gated channel subfamily E/KQT member 1 |
|---|
| Synonyms: | Voltage-gated potassium channel beta subunit Mink/subunit Kv7.1 | Voltage-gated potassium channel, IKs; KCNQ1(Kv7.1)/KCNE1(MinK) |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of EBI is 158552 |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Potassium voltage-gated channel subfamily E member 1 |
|---|
| Synonyms: | Delayed rectifier potassium channel subunit IsK | IKs producing slow voltage-gated potassium channel subunit beta Mink | KCNE1 | KCNE1_HUMAN | KCNQ1(Kv7.1)/KCNE1(MinK) | Minimal potassium channel | Voltage-gated potassium channel beta subunit Mink | Voltage-gated potassium channel, IKs |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 14676.11 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1456351 |
|---|
| Residue: | 129 |
|---|
| Sequence: | MILSNTTAVTPFLTKLWQETVQQGGNMSGLARRSPRSSDGKLEALYVLMVLGFFGFFTLG
IMLSYIRSKKLEHSNDPFNVYIESDAWQEKDKAYVQARVLESYRSCYVVENHLAIEQPNT
HLPETKPSP
|
|
|
|---|
| Component 2 |
| Name: | Potassium voltage-gated channel subfamily KQT member 1 |
|---|
| Synonyms: | KCNA8 | KCNA9 | KCNQ (Kv7) potassium channel | KCNQ1 | KCNQ1_HUMAN | KVLQT1 | Voltage-gated potassium channel subunit Kv7.1 | Voltage-gated potassium channel subunit Kv7.1/Misshapen-like kinase 1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 74739.00 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1454245 |
|---|
| Residue: | 676 |
|---|
| Sequence: | MAAASSPPRAERKRWGWGRLPGARRGSAGLAKKCPFSLELAEGGPAGGALYAPIAPGAPG
PAPPASPAAPAAPPVASDLGPRPPVSLDPRVSIYSTRRPVLARTHVQGRVYNFLERPTGW
KCFVYHFAVFLIVLVCLIFSVLSTIEQYAALATGTLFWMEIVLVVFFGTEYVVRLWSAGC
RSKYVGLWGRLRFARKPISIIDLIVVVASMVVLCVGSKGQVFATSAIRGIRFLQILRMLH
VDRQGGTWRLLGSVVFIHRQELITTLYIGFLGLIFSSYFVYLAEKDAVNESGRVEFGSYA
DALWWGVVTVTTIGYGDKVPQTWVGKTIASCFSVFAISFFALPAGILGSGFALKVQQKQR
QKHFNRQIPAAASLIQTAWRCYAAENPDSSTWKIYIRKAPRSHTLLSPSPKPKKSVVVKK
KKFKLDKDNGVTPGEKMLTVPHITCDPPEERRLDHFSVDGYDSSVRKSPTLLEVSMPHFM
RTNSFAEDLDLEGETLLTPITHISQLREHHRATIKVIRRMQYFVAKKKFQQARKPYDVRD
VIEQYSQGHLNLMVRIKELQRRLDQSIGKPSLFISVSEKSKDRGSNTIGARLNRVEDKVT
QLDQRLALITDMLHQLLSLHGGSTPGSGGPPREGGAHITQPCGSGGSVDPELFLPSNTLP
TYEQLTVPRRGPDEGS
|
|
|
|---|
| BDBM50103642 |
|---|
| n/a |
|---|
| Name | BDBM50103642 |
|---|
| Synonyms: | 4-(6-amino-5-bromo-2-(4-cyanophenylamino)pyrimidin-4-ylamino)-3,5-dimethylbenzonitrile | 4-(6-amino-5-bromo-2-(4-cyanophenylamino)pyrimidin-4-yloxy)-3,5-dimethylbenzonitrile | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE | 4-[6-amino-5-bromo-2-(4-cyano-phenylamino)-pyrimidin-4-yloxy]-3,5-dimethyl-benzonitrile | CHEMBL308954 | ETRAVIRINE | Etravirine (ETR) | Etravirine (ETV) | Etravirine (TMC125) | TMC 125 | TMC-125 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H15BrN6O |
|---|
| Mol. Mass. | 435.277 |
|---|
| SMILES | Cc1cc(cc(C)c1Oc1nc(Nc2ccc(cc2)C#N)nc(N)c1Br)C#N |
|---|
| Structure | 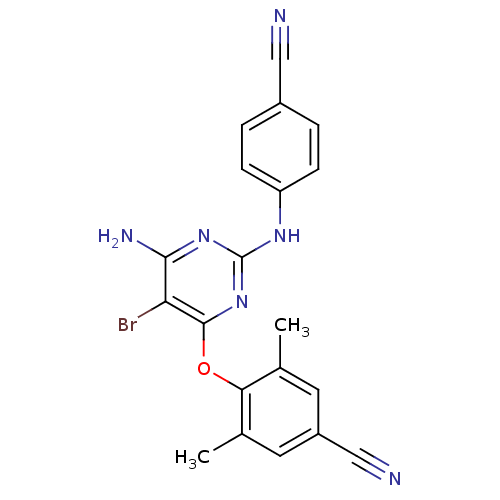
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods70:246-54 (2014) [PubMed] Article
Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods70:246-54 (2014) [PubMed] Article