| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Deoxyribonuclease-1 |
|---|
| Ligand | BDBM50121717 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1516649 (CHEMBL3620429) |
|---|
| IC50 | >100000±n/a nM |
|---|
| Citation |  Chapman, TM; Gillen, KJ; Wallace, C; Lee, MT; Bakrania, P; Khurana, P; Coombs, PJ; Stennett, L; Fox, S; Bureau, EA; Brownlees, J; Melton, DW; Saxty, B Catechols and 3-hydroxypyridones as inhibitors of the DNA repair complex ERCC1-XPF. Bioorg Med Chem Lett25:4097-103 (2015) [PubMed] Article Chapman, TM; Gillen, KJ; Wallace, C; Lee, MT; Bakrania, P; Khurana, P; Coombs, PJ; Stennett, L; Fox, S; Bureau, EA; Brownlees, J; Melton, DW; Saxty, B Catechols and 3-hydroxypyridones as inhibitors of the DNA repair complex ERCC1-XPF. Bioorg Med Chem Lett25:4097-103 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Deoxyribonuclease-1 |
|---|
| Name: | Deoxyribonuclease-1 |
|---|
| Synonyms: | DNAS1_HUMAN | DNASE1 | DNL1 | DNL1 | DNase I | DRNI | Deoxyribonuclease I | Deoxyribonuclease-1 | INN=Dornase alfa |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 31422.78 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_109543 |
|---|
| Residue: | 282 |
|---|
| Sequence: | MRGMKLLGALLALAALLQGAVSLKIAAFNIQTFGETKMSNATLVSYIVQILSRYDIALVQ
EVRDSHLTAVGKLLDNLNQDAPDTYHYVVSEPLGRNSYKERYLFVYRPDQVSAVDSYYYD
DGCEPCGNDTFNREPAIVRFFSRFTEVREFAIVPLHAAPGDAVAEIDALYDVYLDVQEKW
GLEDVMLMGDFNAGCSYVRPSQWSSIRLWTSPTFQWLIPDSADTTATPTHCAYDRIVVAG
MLLRGAVVPDSALPFNFQAAYGLSDQLAQAISDHYPVEVMLK
|
|
|
|---|
| BDBM50121717 |
|---|
| n/a |
|---|
| Name | BDBM50121717 |
|---|
| Synonyms: | CHEMBL3617015 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H12Cl3NO3 |
|---|
| Mol. Mass. | 360.62 |
|---|
| SMILES | Oc1ccc(CNC(=O)Cc2ccc(Cl)cc2Cl)c(Cl)c1O |
|---|
| Structure | 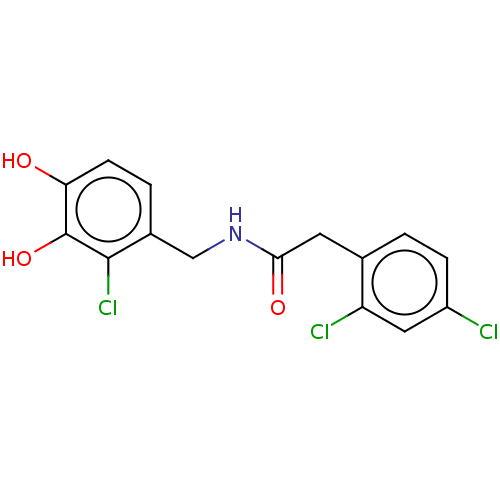
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chapman, TM; Gillen, KJ; Wallace, C; Lee, MT; Bakrania, P; Khurana, P; Coombs, PJ; Stennett, L; Fox, S; Bureau, EA; Brownlees, J; Melton, DW; Saxty, B Catechols and 3-hydroxypyridones as inhibitors of the DNA repair complex ERCC1-XPF. Bioorg Med Chem Lett25:4097-103 (2015) [PubMed] Article
Chapman, TM; Gillen, KJ; Wallace, C; Lee, MT; Bakrania, P; Khurana, P; Coombs, PJ; Stennett, L; Fox, S; Bureau, EA; Brownlees, J; Melton, DW; Saxty, B Catechols and 3-hydroxypyridones as inhibitors of the DNA repair complex ERCC1-XPF. Bioorg Med Chem Lett25:4097-103 (2015) [PubMed] Article