| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Heat shock cognate 71 kDa protein |
|---|
| Ligand | BDBM32375 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1588265 (CHEMBL3825930) |
|---|
| Kd | 720±n/a nM |
|---|
| Citation |  Cheeseman, MD; Westwood, IM; Barbeau, O; Rowlands, M; Dobson, S; Jones, AM; Jeganathan, F; Burke, R; Kadi, N; Workman, P; Collins, I; van Montfort, RL; Jones, K Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of HSP70. J Med Chem59:4625-36 (2016) [PubMed] Article Cheeseman, MD; Westwood, IM; Barbeau, O; Rowlands, M; Dobson, S; Jones, AM; Jeganathan, F; Burke, R; Kadi, N; Workman, P; Collins, I; van Montfort, RL; Jones, K Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of HSP70. J Med Chem59:4625-36 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Heat shock cognate 71 kDa protein |
|---|
| Name: | Heat shock cognate 71 kDa protein |
|---|
| Synonyms: | HSC70 | HSP73 | HSP7C_HUMAN | HSPA10 | HSPA8 | heat shock 70kDa protein 8 isoform 1 | heat shock 70kDa protein 8 isoform 2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 70887.67 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_5729877 |
|---|
| Residue: | 646 |
|---|
| Sequence: | MSKGPAVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVA
MNPTNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVNDAGRPKVQVEYKGETKSFYPEEVS
SMVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAA
IAYGLDKKVGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDFDNRMVNH
FIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEIDSLYEGIDFYTSITRA
RFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLLQDFFNGKELN
KSINPDEAVAYGAAVQAAILSGDKSENVQDLLLLDVTPLSLGIETAGGVMTVLIKRNTTI
PTKQTQTFTTYSDNQPGVLIQVYEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDI
DANGILNVSAVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKN
SLESYAFNMKATVEDEKLQGKINDEDKQKILDKCNEIINWLDKNQTAEKEEFEHQQKELE
KVCNPIITKLYQSAGGMPGGMPGGFPGGGAPPSGGASSGPTIEEVD
|
|
|
|---|
| BDBM32375 |
|---|
| n/a |
|---|
| Name | BDBM32375 |
|---|
| Synonyms: | adenosine-derived inhibitor (Grp78), 9 | adenosine-derived inhibitor, 9 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H21N7O4 |
|---|
| Mol. Mass. | 423.4252 |
|---|
| SMILES | Nc1ncnc2n([C@@H]3O[C@H](CO)[C@@H](O)[C@H]3O)c(NCc3ccc4ncccc4c3)nc12 |r| |
|---|
| Structure | 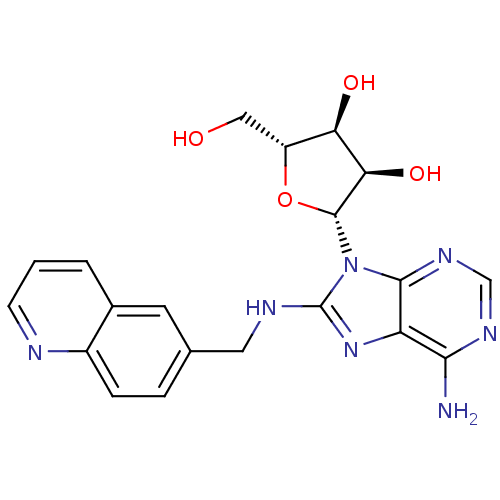
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cheeseman, MD; Westwood, IM; Barbeau, O; Rowlands, M; Dobson, S; Jones, AM; Jeganathan, F; Burke, R; Kadi, N; Workman, P; Collins, I; van Montfort, RL; Jones, K Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of HSP70. J Med Chem59:4625-36 (2016) [PubMed] Article
Cheeseman, MD; Westwood, IM; Barbeau, O; Rowlands, M; Dobson, S; Jones, AM; Jeganathan, F; Burke, R; Kadi, N; Workman, P; Collins, I; van Montfort, RL; Jones, K Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of HSP70. J Med Chem59:4625-36 (2016) [PubMed] Article