| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA gyrase subunit A/B |
|---|
| Ligand | BDBM50215362 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_50467 (CHEMBL658170) |
|---|
| IC50 | >1920000±n/a nM |
|---|
| Citation |  Weinder-Wells, MA; Altom, J; Fernandez, J; Fraga-Spano, SA; Hilliard, J; Ohemeng, K; Barrett, JF DNA gyrase inhibitory activity of ellagic acid derivatives. Bioorg Med Chem Lett8:97-100 (1998) [PubMed] Weinder-Wells, MA; Altom, J; Fernandez, J; Fraga-Spano, SA; Hilliard, J; Ohemeng, K; Barrett, JF DNA gyrase inhibitory activity of ellagic acid derivatives. Bioorg Med Chem Lett8:97-100 (1998) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA gyrase subunit A/B |
|---|
| Name: | DNA gyrase subunit A/B |
|---|
| Synonyms: | DNA Gyrase | DNA gyrase A/B |
|---|
| Type: | A2B2 tetramer |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | DNA gyrase subunit A |
|---|
| Synonyms: | DNA gyrase | DNA gyrase subunit A (gyrA) | GYRA_ECOLI | gyrA | hisW | nalA | parD |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 96935.15 |
|---|
| Organism: | Escherichia coli (strain K12) |
|---|
| Description: | n/a |
|---|
| Residue: | 875 |
|---|
| Sequence: | MSDLAREITPVNIEEELKSSYLDYAMSVIVGRALPDVRDGLKPVHRRVLYAMNVLGNDWN
KAYKKSARVVGDVIGKYHPHGDSAVYDTIVRMAQPFSLRYMLVDGQGNFGSIDGDSAAAM
RYTEIRLAKIAHELMADLEKETVDFVDNYDGTEKIPDVMPTKIPNLLVNGSSGIAVGMAT
NIPPHNLTEVINGCLAYIDDEDISIEGLMEHIPGPDFPTAAIINGRRGIEEAYRTGRGKV
YIRARAEVEVDAKTGRETIIVHEIPYQVNKARLIEKIAELVKEKRVEGISALRDESDKDG
MRIVIEVKRDAVGEVVLNNLYSQTQLQVSFGINMVALHHGQPKIMNLKDIIAAFVRHRRE
VVTRRTIFELRKARDRAHILEALAVALANIDPIIELIRHAPTPAEAKTALVANPWQLGNV
AAMLERAGDDAARPEWLEPEFGVRDGLYYLTEQQAQAILDLRLQKLTGLEHEKLLDEYKE
LLDQIAELLRILGSADRLMEVIREELELVREQFGDKRRTEITANSADINLEDLITQEDVV
VTLSHQGYVKYQPLSEYEAQRRGGKGKSAARIKEEDFIDRLLVANTHDHILCFSSRGRVY
SMKVYQLPEATRGARGRPIVNLLPLEQDERITAILPVTEFEEGVKVFMATANGTVKKTVL
TEFNRLRTAGKVAIKLVDGDELIGVDLTSGEDEVMLFSAEGKVVRFKESSVRAMGCNTTG
VRGIRLGEGDKVVSLIVPRGDGAILTATQNGYGKRTAVAEYPTKSRATKGVISIKVTERN
GLVVGAVQVDDCDQIMMITDAGTLVRTRVSEISIVGRNTQGVILIRTAEDENVVGLQRVA
EPVDEEDLDTIDGSAAEGDDEIAPEVDVDDEPEEE
|
|
|
|---|
| Component 2 |
| Name: | DNA gyrase subunit B |
|---|
| Synonyms: | DNA gyrase subunit B | DNA gyrase subunit B (gyrB) | GYRB_ECOLI | Type IIA topoisomerase subunit GyrB | acrB | cou | gyrB | himB | hisU | nalC | parA | pcbA |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 89941.28 |
|---|
| Organism: | Escherichia coli (strain K12) |
|---|
| Description: | P0AES6 |
|---|
| Residue: | 804 |
|---|
| Sequence: | MSNSYDSSSIKVLKGLDAVRKRPGMYIGDTDDGTGLHHMVFEVVDNAIDEALAGHCKEII
VTIHADNSVSVQDDGRGIPTGIHPEEGVSAAEVIMTVLHAGGKFDDNSYKVSGGLHGVGV
SVVNALSQKLELVIQREGKIHRQIYEHGVPQAPLAVTGETEKTGTMVRFWPSLETFTNVT
EFEYEILAKRLRELSFLNSGVSIRLRDKRDGKEDHFHYEGGIKAFVEYLNKNKTPIHPNI
FYFSTEKDGIGVEVALQWNDGFQENIYCFTNNIPQRDGGTHLAGFRAAMTRTLNAYMDKE
GYSKKAKVSATGDDAREGLIAVVSVKVPDPKFSSQTKDKLVSSEVKSAVEQQMNELLAEY
LLENPTDAKIVVGKIIDAARAREAARRAREMTRRKGALDLAGLPGKLADCQERDPALSEL
YLVEGDSAGGSAKQGRNRKNQAILPLKGKILNVEKARFDKMLSSQEVATLITALGCGIGR
DEYNPDKLRYHSIIIMTDADVDGSHIRTLLLTFFYRQMPEIVERGHVYIAQPPLYKVKKG
KQEQYIKDDEAMDQYQISIALDGATLHTNASAPALAGEALEKLVSEYNATQKMINRMERR
YPKAMLKELIYQPTLTEADLSDEQTVTRWVNALVSELNDKEQHGSQWKFDVHTNAEQNLF
EPIVRVRTHGVDTDYPLDHEFITGGEYRRICTLGEKLRGLLEEDAFIERGERRQPVASFE
QALDWLVKESRRGLSIQRYKGLGEMNPEQLWETTMDPESRRMLRVTVKDAIAADQLFTTL
MGDAVEPRRAFIEENALKAANIDI
|
|
|
|---|
| BDBM50215362 |
|---|
| n/a |
|---|
| Name | BDBM50215362 |
|---|
| Synonyms: | CHEMBL6336 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H9FO4 |
|---|
| Mol. Mass. | 260.2173 |
|---|
| SMILES | Cc1cc(O)c(O)c2oc(=O)c3ccc(F)cc3c12 |
|---|
| Structure | 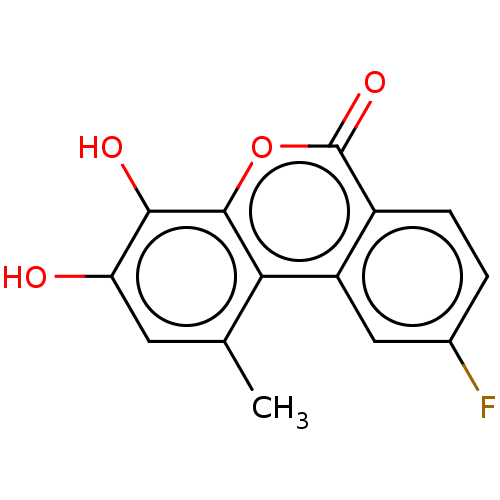
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Weinder-Wells, MA; Altom, J; Fernandez, J; Fraga-Spano, SA; Hilliard, J; Ohemeng, K; Barrett, JF DNA gyrase inhibitory activity of ellagic acid derivatives. Bioorg Med Chem Lett8:97-100 (1998) [PubMed]
Weinder-Wells, MA; Altom, J; Fernandez, J; Fraga-Spano, SA; Hilliard, J; Ohemeng, K; Barrett, JF DNA gyrase inhibitory activity of ellagic acid derivatives. Bioorg Med Chem Lett8:97-100 (1998) [PubMed]