

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Type-1 angiotensin II receptor A/Type-1 angiotensin II receptor B/Type-2 angiotensin II receptor | ||
| Ligand | BDBM50228204 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_36479 (CHEMBL651548) | ||
| Ki | 1.2±n/a nM | ||
| Citation |  Hsieh, KH; LaHann, TR; Speth, RC Topographic probes of angiotensin and receptor: potent angiotensin II agonist containing diphenylalanine and long-acting antagonists containing biphenylalanine and 2-indan amino acid in position 8. J Med Chem32:898-903 (1989) [PubMed] Hsieh, KH; LaHann, TR; Speth, RC Topographic probes of angiotensin and receptor: potent angiotensin II agonist containing diphenylalanine and long-acting antagonists containing biphenylalanine and 2-indan amino acid in position 8. J Med Chem32:898-903 (1989) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Type-1 angiotensin II receptor A/Type-1 angiotensin II receptor B/Type-2 angiotensin II receptor | |||
| Name: | Type-1 angiotensin II receptor A/Type-1 angiotensin II receptor B/Type-2 angiotensin II receptor | ||
| Synonyms: | Angiotensin II receptor | Type-1A/Type-1B/Type-2 angiotensin II receptor | ||
| Type: | n/a | ||
| Mol. Mass.: | n/a | ||
| Description: | ASSAY_ID of ChEMBL is 36477 | ||
| Components: | This complex has 3 components. | ||
| Component 1 | |||
| Name: | Type-1 angiotensin II receptor B | ||
| Synonyms: | AGTRB_RAT | AT3 | Agtr1 | Agtr1b | Angiotensin II AT1B | Angiotensin II receptor (AT-1) type-1 | Angiotensin II type 1b (AT-1b) receptor | At1b | Type-1B angiotensin II receptor | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 40929.44 | ||
| Organism: | RAT | ||
| Description: | Angiotensin II AT1B 0 RAT::P29089 | ||
| Residue: | 359 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | Type-2 angiotensin II receptor | ||
| Synonyms: | AGTR2_RAT | AT2 | Agtr2 | Angiotensin II AT2 | Angiotensin II receptor | Angiotensin II type 2 (AT-2) receptor | Type-2 angiotensin II receptor | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 41346.62 | ||
| Organism: | RAT | ||
| Description: | Angiotensin II AT2 0 0::P35351 | ||
| Residue: | 363 | ||
| Sequence: |
| ||
| Component 3 | |||
| Name: | Type-1 angiotensin II receptor A | ||
| Synonyms: | AGTRA_RAT | ANGIOTENSIN AT1 | Agtr1 | Agtr1a | Angiotensin II AT1 | Angiotensin II AT1A | Angiotensin II receptor (AT-1) type-1 | At1a | Type-1A angiotensin II receptor | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 40910.53 | ||
| Organism: | RAT | ||
| Description: | ANGIOTENSIN AT1 AGTR1 RAT::P25095 | ||
| Residue: | 359 | ||
| Sequence: |
| ||
| BDBM50228204 | |||
| n/a | |||
| Name | BDBM50228204 | ||
| Synonyms: | CHEMBL405389 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C54H73N13O10 | ||
| Mol. Mass. | 1064.2385 | ||
| SMILES | CNCC(=O)N[C@@H](CCCN=C(N)N)C(=O)N[C@@H](C(C)C)C(=O)N[C@@H](Cc1ccc(O)cc1)C(=O)N[C@@H](C(C)C)C(=O)N[C@@H](Cc1c[nH]cn1)C(=O)N1CCC[C@@H]1C(=O)N[C@@H](C(c1ccccc1)c1ccccc1)C(O)=O |wU:56.59,36.36,17.16,wD:60.62,43.43,24.23,6.5,(2.9,-15.86,;2.9,-14.33,;4.23,-13.56,;5.56,-14.33,;5.56,-15.86,;6.9,-13.56,;8.24,-14.33,;8.24,-15.86,;9.56,-16.64,;9.56,-18.18,;10.91,-18.95,;10.91,-20.49,;12.24,-21.26,;9.56,-21.26,;9.56,-13.56,;9.56,-12.02,;10.91,-14.33,;12.24,-13.56,;12.24,-12.02,;13.57,-11.23,;10.91,-11.23,;13.57,-14.33,;13.57,-15.86,;14.91,-13.56,;16.25,-14.33,;16.25,-15.86,;17.57,-16.64,;17.57,-18.18,;18.92,-18.95,;20.25,-18.18,;21.58,-18.95,;20.25,-16.64,;18.92,-15.86,;17.57,-13.56,;17.57,-12.02,;18.92,-14.33,;20.25,-13.56,;20.25,-12.02,;21.58,-11.23,;18.92,-11.23,;21.58,-14.33,;21.58,-15.86,;22.93,-13.56,;24.26,-14.33,;24.26,-15.86,;24.82,-17.35,;24.98,-18.88,;26.48,-19.2,;27.27,-17.86,;26.23,-16.71,;25.59,-13.56,;25.59,-12.02,;26.93,-14.33,;27.09,-15.86,;28.6,-16.18,;29.37,-14.85,;28.34,-13.7,;28.66,-12.19,;30.13,-11.71,;27.51,-11.16,;27.85,-9.66,;29.3,-9.18,;29.62,-7.67,;31.09,-7.19,;31.41,-5.68,;30.26,-4.65,;28.79,-5.13,;28.47,-6.63,;30.84,-9.18,;31.6,-10.49,;33.14,-10.49,;33.89,-9.17,;33.12,-7.83,;31.58,-7.85,;26.7,-8.62,;27.02,-7.11,;25.23,-9.1,)| | ||
| Structure | 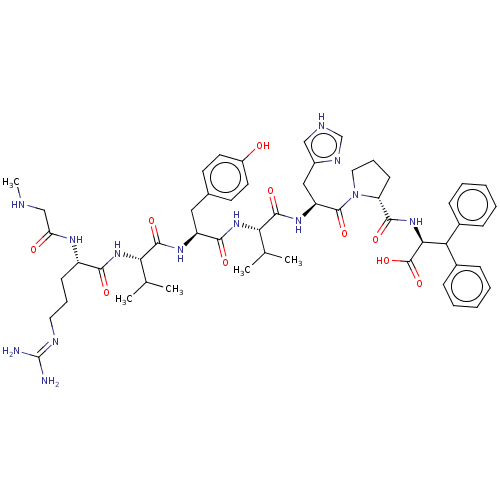
| ||