

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50237503 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1660136 (CHEMBL4009748) | ||
| IC50 | 780±n/a nM | ||
| Citation |  Wu, YJ; Guernon, J; Shi, J; Ditta, J; Robbins, KJ; Rajamani, R; Easton, A; Newton, A; Bourin, C; Mosure, K; Soars, MG; Knox, RJ; Matchett, M; Pieschl, RL; Post-Munson, DJ; Wang, S; Herrington, J; Graef, J; Newberry, K; Bristow, LJ; Meanwell, NA; Olson, R; Thompson, LA; Dzierba, C Development of New Benzenesulfonamides As Potent and Selective Na J Med Chem60:2513-2525 (2017) [PubMed] Article Wu, YJ; Guernon, J; Shi, J; Ditta, J; Robbins, KJ; Rajamani, R; Easton, A; Newton, A; Bourin, C; Mosure, K; Soars, MG; Knox, RJ; Matchett, M; Pieschl, RL; Post-Munson, DJ; Wang, S; Herrington, J; Graef, J; Newberry, K; Bristow, LJ; Meanwell, NA; Olson, R; Thompson, LA; Dzierba, C Development of New Benzenesulfonamides As Potent and Selective Na J Med Chem60:2513-2525 (2017) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50237503 | |||
| n/a | |||
| Name | BDBM50237503 | ||
| Synonyms: | CHEMBL4093720 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H22F2N4O3S2 | ||
| Mol. Mass. | 432.508 | ||
| SMILES | C[C@@]1(CCOc2cc(F)c(cc2F)S(=O)(=O)Nc2ncns2)CC[C@H](N)CC1 |r,wU:1.1,24.26,(59.23,-38.27,;60.33,-39.38,;61.11,-38.05,;62.65,-38.05,;63.41,-39.39,;64.95,-39.4,;65.72,-38.08,;67.25,-38.08,;68.03,-36.75,;68.03,-39.42,;67.25,-40.75,;65.71,-40.74,;64.93,-42.07,;69.56,-39.42,;69.98,-37.93,;71.06,-39.04,;70.33,-40.76,;71.87,-40.77,;72.78,-39.53,;74.24,-40.01,;74.24,-41.55,;72.77,-42.02,;58.98,-40.09,;58.93,-41.63,;60.24,-42.44,;60.19,-43.98,;61.6,-41.72,;61.65,-40.17,)| | ||
| Structure | 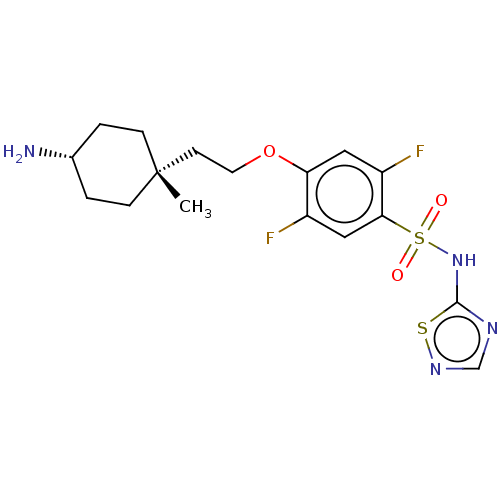
| ||