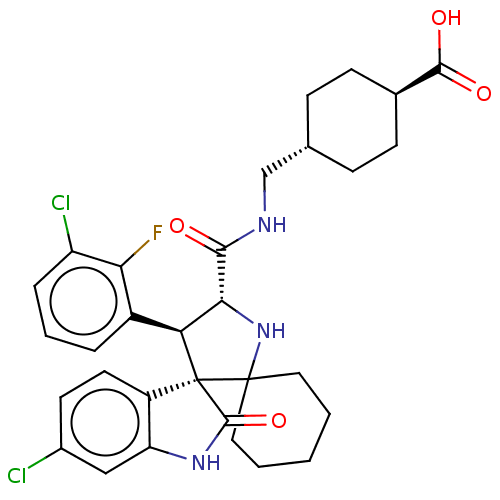
| Citation |  Aguilar, A; Lu, J; Liu, L; Du, D; Bernard, D; McEachern, D; Przybranowski, S; Li, X; Luo, R; Wen, B; Sun, D; Wang, H; Wen, J; Wang, G; Zhai, Y; Guo, M; Yang, D; Wang, S Discovery of 4-((3'R,4'S,5'R)-6¿-Chloro-4'-(3-chloro-2-fluorophenyl)-1'-ethyl-2¿-oxodispiro[cyclohexane-1,2'-pyrrolidine-3',3¿-indoline]-5'-carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): A Potent and Orally Active Murine Double Minute 2 (MDM2) Inhibitor in Clinical Deve J Med Chem60:2819-2839 (2017) [PubMed] Article Aguilar, A; Lu, J; Liu, L; Du, D; Bernard, D; McEachern, D; Przybranowski, S; Li, X; Luo, R; Wen, B; Sun, D; Wang, H; Wen, J; Wang, G; Zhai, Y; Guo, M; Yang, D; Wang, S Discovery of 4-((3'R,4'S,5'R)-6¿-Chloro-4'-(3-chloro-2-fluorophenyl)-1'-ethyl-2¿-oxodispiro[cyclohexane-1,2'-pyrrolidine-3',3¿-indoline]-5'-carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): A Potent and Orally Active Murine Double Minute 2 (MDM2) Inhibitor in Clinical Deve J Med Chem60:2819-2839 (2017) [PubMed] Article |
|---|
| SMILES | OC(=O)[C@H]1CC[C@H](CNC(=O)[C@@H]2NC3(CCCCC3)[C@]3([C@H]2c2cccc(Cl)c2F)C(=O)Nc2cc(Cl)ccc32)CC1 |r,wU:19.31,20.22,6.6,wD:11.10,3.2,(34.1,-31.43,;32.64,-30.93,;32.34,-29.42,;31.48,-31.94,;30.02,-31.45,;28.86,-32.46,;29.17,-33.97,;28.02,-34.98,;26.56,-34.49,;25.4,-35.51,;23.97,-34.95,;25.78,-37,;27.22,-37.55,;27.15,-39.08,;27.54,-40.57,;29.02,-40.96,;30.11,-39.88,;29.71,-38.4,;28.23,-37.99,;25.66,-39.49,;24.73,-38.23,;23.39,-36.71,;23.09,-35.19,;21.63,-34.7,;20.47,-35.71,;20.77,-37.22,;19.6,-38.23,;22.22,-37.72,;23.91,-38.71,;26.58,-40.74,;27.98,-41.9,;25.67,-42,;24.2,-41.53,;22.86,-42.3,;21.52,-41.53,;20.19,-42.3,;21.52,-39.99,;22.85,-39.22,;24.19,-39.98,;30.63,-34.46,;31.78,-33.45,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Aguilar, A; Lu, J; Liu, L; Du, D; Bernard, D; McEachern, D; Przybranowski, S; Li, X; Luo, R; Wen, B; Sun, D; Wang, H; Wen, J; Wang, G; Zhai, Y; Guo, M; Yang, D; Wang, S Discovery of 4-((3'R,4'S,5'R)-6¿-Chloro-4'-(3-chloro-2-fluorophenyl)-1'-ethyl-2¿-oxodispiro[cyclohexane-1,2'-pyrrolidine-3',3¿-indoline]-5'-carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): A Potent and Orally Active Murine Double Minute 2 (MDM2) Inhibitor in Clinical Deve J Med Chem60:2819-2839 (2017) [PubMed] Article
Aguilar, A; Lu, J; Liu, L; Du, D; Bernard, D; McEachern, D; Przybranowski, S; Li, X; Luo, R; Wen, B; Sun, D; Wang, H; Wen, J; Wang, G; Zhai, Y; Guo, M; Yang, D; Wang, S Discovery of 4-((3'R,4'S,5'R)-6¿-Chloro-4'-(3-chloro-2-fluorophenyl)-1'-ethyl-2¿-oxodispiro[cyclohexane-1,2'-pyrrolidine-3',3¿-indoline]-5'-carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): A Potent and Orally Active Murine Double Minute 2 (MDM2) Inhibitor in Clinical Deve J Med Chem60:2819-2839 (2017) [PubMed] Article