| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Myeloperoxidase |
|---|
| Ligand | BDBM50238832 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1664234 (CHEMBL4013915) |
|---|
| IC50 | 40±n/a nM |
|---|
| Citation |  Soubhye, J; Chikh Alard, I; Aldib, I; Prévost, M; Gelbcke, M; De Carvalho, A; Furtmüller, PG; Obinger, C; Flemmig, J; Tadrent, S; Meyer, F; Rousseau, A; Nève, J; Mathieu, V; Zouaoui Boudjeltia, K; Dufrasne, F; Van Antwerpen, P Discovery of Novel Potent Reversible and Irreversible Myeloperoxidase Inhibitors Using Virtual Screening Procedure. J Med Chem60:6563-6586 (2017) [PubMed] Article Soubhye, J; Chikh Alard, I; Aldib, I; Prévost, M; Gelbcke, M; De Carvalho, A; Furtmüller, PG; Obinger, C; Flemmig, J; Tadrent, S; Meyer, F; Rousseau, A; Nève, J; Mathieu, V; Zouaoui Boudjeltia, K; Dufrasne, F; Van Antwerpen, P Discovery of Novel Potent Reversible and Irreversible Myeloperoxidase Inhibitors Using Virtual Screening Procedure. J Med Chem60:6563-6586 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Myeloperoxidase |
|---|
| Name: | Myeloperoxidase |
|---|
| Synonyms: | 84 kDa myeloperoxidase | 89 kDa myeloperoxidase | MPO | Mpo protein | Myeloperoxidase | Myeloperoxidase (MPO) | Myeloperoxidase heavy chain | Myeloperoxidase light chain | PERM_HUMAN |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 83888.32 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P05164 |
|---|
| Residue: | 745 |
|---|
| Sequence: | MGVPFFSSLRCMVDLGPCWAGGLTAEMKLLLALAGLLAILATPQPSEGAAPAVLGEVDTS
LVLSSMEEAKQLVDKAYKERRESIKQRLRSGSASPMELLSYFKQPVAATRTAVRAADYLH
VALDLLERKLRSLWRRPFNVTDVLTPAQLNVLSKSSGCAYQDVGVTCPEQDKYRTITGMC
NNRRSPTLGASNRAFVRWLPAEYEDGFSLPYGWTPGVKRNGFPVALARAVSNEIVRFPTD
QLTPDQERSLMFMQWGQLLDHDLDFTPEPAARASFVTGVNCETSCVQQPPCFPLKIPPND
PRIKNQADCIPFFRSCPACPGSNITIRNQINALTSFVDASMVYGSEEPLARNLRNMSNQL
GLLAVNQRFQDNGRALLPFDNLHDDPCLLTNRSARIPCFLAGDTRSSEMPELTSMHTLLL
REHNRLATELKSLNPRWDGERLYQEARKIVGAMVQIITYRDYLPLVLGPTAMRKYLPTYR
SYNDSVDPRIANVFTNAFRYGHTLIQPFMFRLDNRYQPMEPNPRVPLSRVFFASWRVVLE
GGIDPILRGLMATPAKLNRQNQIAVDEIRERLFEQVMRIGLDLPALNMQRSRDHGLPGYN
AWRRFCGLPQPETVGQLGTVLRNLKLARKLMEQYGTPNNIDIWMGGVSEPLKRKGRVGPL
LACIIGTQFRKLRDGDRFWWENEGVFSMQQRQALAQISLPRIICDNTGITTVSKNNIFMS
NSYPRDFVNCSTLPALNLASWREAS
|
|
|
|---|
| BDBM50238832 |
|---|
| n/a |
|---|
| Name | BDBM50238832 |
|---|
| Synonyms: | CHEMBL4080413 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H10FN5 |
|---|
| Mol. Mass. | 219.2183 |
|---|
| SMILES | [#6]-c1nc(\[#7]=[#6](/[#7])-[#7])nc2cc(F)ccc12 |
|---|
| Structure | 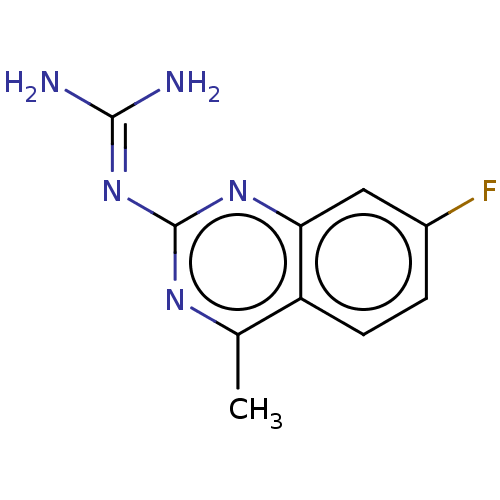
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Soubhye, J; Chikh Alard, I; Aldib, I; Prévost, M; Gelbcke, M; De Carvalho, A; Furtmüller, PG; Obinger, C; Flemmig, J; Tadrent, S; Meyer, F; Rousseau, A; Nève, J; Mathieu, V; Zouaoui Boudjeltia, K; Dufrasne, F; Van Antwerpen, P Discovery of Novel Potent Reversible and Irreversible Myeloperoxidase Inhibitors Using Virtual Screening Procedure. J Med Chem60:6563-6586 (2017) [PubMed] Article
Soubhye, J; Chikh Alard, I; Aldib, I; Prévost, M; Gelbcke, M; De Carvalho, A; Furtmüller, PG; Obinger, C; Flemmig, J; Tadrent, S; Meyer, F; Rousseau, A; Nève, J; Mathieu, V; Zouaoui Boudjeltia, K; Dufrasne, F; Van Antwerpen, P Discovery of Novel Potent Reversible and Irreversible Myeloperoxidase Inhibitors Using Virtual Screening Procedure. J Med Chem60:6563-6586 (2017) [PubMed] Article