

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | N-formyl peptide receptor 2 | ||
| Ligand | BDBM50238898 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1664517 (CHEMBL4014198) | ||
| IC50 | 273±n/a nM | ||
| Citation |  Skovbakke, SL; Skovbakke, SL; Skovbakke, SL; Holdfeldt, A; Holdfeldt, A; Holdfeldt, A; Nielsen, C; Nielsen, C; Nielsen, C; Hansen, AM; Hansen, AM; Hansen, AM; Perez-Gassol, I; Perez-Gassol, I; Perez-Gassol, I; Dahlgren, C; Dahlgren, C; Dahlgren, C; Forsman, H; Forsman, H; Forsman, H; Franzyk, H; Franzyk, H; Franzyk, H Combining Elements from Two Antagonists of Formyl Peptide Receptor 2 Generates More Potent Peptidomimetic Antagonists. J Med Chem60:6991-6997 (2017) [PubMed] Article Skovbakke, SL; Skovbakke, SL; Skovbakke, SL; Holdfeldt, A; Holdfeldt, A; Holdfeldt, A; Nielsen, C; Nielsen, C; Nielsen, C; Hansen, AM; Hansen, AM; Hansen, AM; Perez-Gassol, I; Perez-Gassol, I; Perez-Gassol, I; Dahlgren, C; Dahlgren, C; Dahlgren, C; Forsman, H; Forsman, H; Forsman, H; Franzyk, H; Franzyk, H; Franzyk, H Combining Elements from Two Antagonists of Formyl Peptide Receptor 2 Generates More Potent Peptidomimetic Antagonists. J Med Chem60:6991-6997 (2017) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| N-formyl peptide receptor 2 | |||
| Name: | N-formyl peptide receptor 2 | ||
| Synonyms: | ALXR, FPRL1, FPR2 | FMLP-related receptor I FMLP-R-I | FPR2 | FPR2_HUMAN | FPRH1 | FPRL1 | Formyl Peptide Receptor-Like 1 | HM63 | LXA4 receptor | LXA4R | Lipoxin A4 receptor | Lipoxin A4 receptor (LXA4) | RFP | hFPRL | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 38968.35 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P25090 | ||
| Residue: | 351 | ||
| Sequence: |
| ||
| BDBM50238898 | |||
| n/a | |||
| Name | BDBM50238898 | ||
| Synonyms: | CHEMBL4094101 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C60H78N9O6 | ||
| Mol. Mass. | 1021.3175 | ||
| SMILES | CCN(CC)c1ccc2c(-c3ccccc3C(=O)N[C@@H](CCCCN)C(=O)N(CCC(=O)N[C@@H](CCCCN)C(=O)N(CCC(N)=O)Cc3ccccc3)Cc3ccccc3)c3ccc(cc3oc2c1)=[N+](CC)CC |r,wU:33.34,wD:19.20,(20.03,-41.29,;18.7,-40.52,;18.7,-38.97,;20.03,-38.2,;21.36,-38.97,;17.35,-38.2,;17.34,-36.68,;16.01,-35.91,;14.69,-36.69,;13.35,-35.92,;13.34,-33.85,;11.99,-33.09,;11.99,-31.55,;13.32,-30.77,;14.66,-31.54,;14.66,-33.08,;15.99,-33.85,;15.99,-35.4,;17.34,-33.08,;18.7,-33.85,;18.7,-35.4,;20.03,-36.17,;20.03,-37.71,;21.36,-38.48,;22.7,-37.71,;20.05,-33.08,;20.05,-31.52,;21.39,-33.85,;21.39,-35.39,;22.73,-36.15,;24.06,-35.38,;24.06,-33.84,;25.4,-36.15,;26.73,-35.37,;26.72,-33.83,;28.05,-33.06,;28.05,-31.52,;29.38,-30.75,;29.38,-29.21,;28.06,-36.14,;28.07,-37.68,;29.39,-35.37,;30.73,-36.13,;30.73,-37.67,;32.07,-38.44,;32.07,-39.98,;33.4,-37.67,;29.39,-33.83,;30.72,-33.05,;32.05,-33.82,;33.38,-33.05,;33.38,-31.51,;32.04,-30.74,;30.71,-31.52,;22.72,-33.07,;22.72,-31.53,;24.05,-30.77,;24.05,-29.23,;22.71,-28.46,;21.38,-29.24,;21.39,-30.78,;12.01,-36.7,;10.68,-35.94,;9.36,-36.72,;9.38,-38.26,;10.7,-39.02,;12.03,-38.24,;13.37,-39,;14.69,-38.23,;16.03,-38.99,;8.03,-39.03,;6.69,-38.26,;5.36,-39.03,;8.03,-40.57,;6.69,-41.34,)| | ||
| Structure | 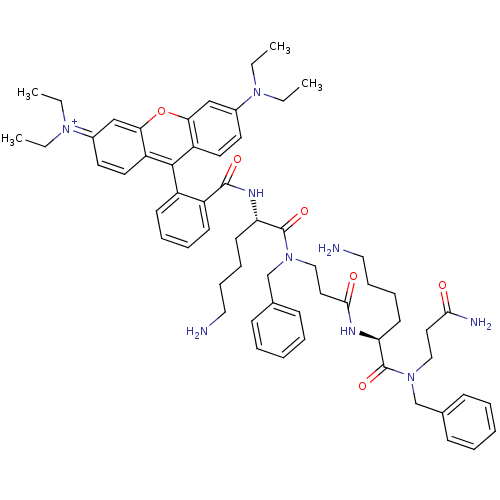
| ||