| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA gyrase subunit A/B |
|---|
| Ligand | BDBM50226181 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1664693 (CHEMBL4014489) |
|---|
| IC50 | 41±n/a nM |
|---|
| Citation |  Jakopin, ?; Ila?, J; Baran?oková, M; Brvar, M; Tammela, P; Sollner Dolenc, M; Toma?i?, T; Kikelj, D Discovery of substituted oxadiazoles as a novel scaffold for DNA gyrase inhibitors. Eur J Med Chem130:171-184 (2017) [PubMed] Article Jakopin, ?; Ila?, J; Baran?oková, M; Brvar, M; Tammela, P; Sollner Dolenc, M; Toma?i?, T; Kikelj, D Discovery of substituted oxadiazoles as a novel scaffold for DNA gyrase inhibitors. Eur J Med Chem130:171-184 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA gyrase subunit A/B |
|---|
| Name: | DNA gyrase subunit A/B |
|---|
| Synonyms: | DNA Gyrase |
|---|
| Type: | A2B2 tetramer |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | DNA gyrase subunit A |
|---|
| Synonyms: | DNA gyrase subunit A (gyrA) | GYRA_STAAU | gyrA |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 99588.82 |
|---|
| Organism: | Staphylococcus aureus |
|---|
| Description: | n/a |
|---|
| Residue: | 889 |
|---|
| Sequence: | MAELPQSRINERNITSEMRESFLDYAMSVIVARALPDVRDGLKPVHRRILYGLNEQGMTP
DKSYKKSARIVGDVMGKYHPHGDSSIYEAMVRMAQDFSYRYPLVDGQGNFGSMDGDGAAA
MRYTEARMTKITLELLRDINKDTIDFIDNYDGNEREPSVLPARFPNLLANGASGIAVGMA
TNIPPHNLTELINGVLSLSKNPDISIAELMEDIEGPDFPTAGLILGKSGIRRAYETGRGS
IQMRSRAVIEERGGGRQRIVVTEIPFQVNKARMIEKIAELVRDKKIDGITDLRDETSLRT
GVRVVIDVRKDANASVILNNLYKQTPLQTSFGVNMIALVNGRPKLINLKEALVHYLEHQK
TVVRRRTQYNLRKAKDRAHILEGLRIALDHIDEIISTIRESDTDKVAMESLQQRFKLSEK
QAQAILDMRLRRLTGLERDKIEAEYNELLNYISELEAILADEEVLLQLVRDELTEIRDRF
GDDRRTEIQLGGFEDLEDEDLIPEEQIVITLSHNNYIKRLPVSTYRAQNRGGRGVQGMNT
LEEDFVSQLVTLSTHDHVLFFTNKGRVYKLKGYEVPELSRQSKGIPVVNAIELENDEVIS
TMIAVKDLESEDNFLVFATKRGVVKRSALSNFSRINRNGKIAISFREDDELIAVRLTSGQ
EDILIGTSHASLIRFPESTLRPLGRTATGVKGITLREGDEVVGLDVAHANSVDEVLVVTE
NGYGKRTPVNDYRLSNRGGKGIKTATITERNGNVVCITTVTGEEDLMIVTNAGVIIRLDV
ADISQNGRAAQGVRLIRLGDDQFVSTVAKVKEDAEDETNEDEQSTSTVSEDGTEQQREAV
VNDETPGNAIHTEVIDSEENDEDGRIEVRQDFMDRVEEDIQQSLDEDEE
|
|
|
|---|
| Component 2 |
| Name: | DNA gyrase subunit B |
|---|
| Synonyms: | DNA gyrase | DNA gyrase subunit B (DNA gyraseB) | DNA gyrase subunit B (gyrB) | GYRB_STAAU | gyrB |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 72530.91 |
|---|
| Organism: | Staphylococcus aureus |
|---|
| Description: | n/a |
|---|
| Residue: | 644 |
|---|
| Sequence: | MVTALSDVNNTDNYGAGQIQVLEGLEAVRKRPGMYIGSTSERGLHHLVWEIVDNSIDEAL
AGYANQIEVVIEKDNWIKVTDNGRGIPVDIQEKMGRPAVEVILTVLHAGGKFGGGGYKVS
GGLHGVGSSVVNALSQDLEVYVHRNETIYHQAYKKGVPQFDLKEVGTTDKTGTVIRFKAD
GEIFTETTVYNYETLQQRIRELAFLNKGIQITLRDERDEENVREDSYHYEGGIKSYVELL
NENKEPIHDEPIYIHQSKDDIEVEIAIQYNSGYATNLLTYANNIHTYEGGTHEDGFKRAL
TRVLNSYGLSSKIMKEEKDRLSGEDTREGMTAIISIKHGDPQFEGQTKTKLGNSEVRQVV
DKLFSEHFERFLYENPQVARTVVEKGIMAARARVAAKKAREVTRRKSALDVASLPGKLAD
CSSKSPEECEIFLVEGDSAGGSTKSGRDSRTQAILPLRGKILNVEKARLDRILNNNEIRQ
MITAFGTGIGGDFDLAKARYHKIVIMTDADVDGAHIRTLLLTFFYRFMRPLIEAGYVYIA
QPPLYKLTQGKQKYYVYNDRELDKLKSELNPTPKWSIARYKGLGEMNADQLWETTMNPEH
RALLQVKLEDAIEADQTFEMLMGDVVENRRQFIEDNAVYANLDF
|
|
|
|---|
| BDBM50226181 |
|---|
| n/a |
|---|
| Name | BDBM50226181 |
|---|
| Synonyms: | (3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-3-(3-methylbut-2-enyl)benzamido)-2-oxo-2H-chromen-7-yloxy)-3-methoxy-2,2-dimethyl-tetrahydro-2H-pyran-4-yl carbamate | (3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-3-(3-methylbut-2-enyl)benzamido)-8-methyl-2-oxo-2H-chromen-7-yloxy)-3-methoxy-2,2-dimethyl-tetrahydro-2H-pyran-4-yl carbamate | CHEMBL36506 | Carbamic acid (3R,4S,5R,6R)-5-hydroxy-6-{4-hydroxy-3-[4-hydroxy-3-(3-methyl-but-2-enyl)-benzoylamino]-8-methyl-2-oxo-2H-chromen-7-yloxy}-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester | Carbamic acid (3R,4S,5R,6R)-5-hydroxy-6-{4-hydroxy-3-[4-hydroxy-3-(4-methyl-pent-3-enyl)-benzoylamino]-8-methyl-2-oxo-2H-chromen-7-yloxy}-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester | Carbamic acid 5-hydroxy-6-{4-hydroxy-3-[4-hydroxy-3-(3-methyl-but-2-enyl)-benzoylamino]-8-methyl-2-oxo-2H-chromen-7-yloxy}-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester | Carbamic acid 5-hydroxy-6-{4-hydroxy-3-[4-hydroxy-3-(3-methyl-but-2-enyl)-benzoylamino]-8-methyl-2-oxo-2H-chromen-7-yloxy}-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester(Novobiocin) | NOVOBIOCIN |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H36N2O11 |
|---|
| Mol. Mass. | 612.6243 |
|---|
| SMILES | [#6]-[#8]-[#6@@H]1-[#6@@H](-[#8]-[#6](-[#7])=O)-[#6@@H](-[#8])-[#6@H](-[#8]-c2ccc3c(-[#8])c(-[#7]-[#6](=O)-c4ccc(-[#8])c(-[#6]\[#6]=[#6](/[#6])-[#6])c4)c(=O)oc3c2-[#6])-[#8]C1([#6])[#6] |r| |
|---|
| Structure | 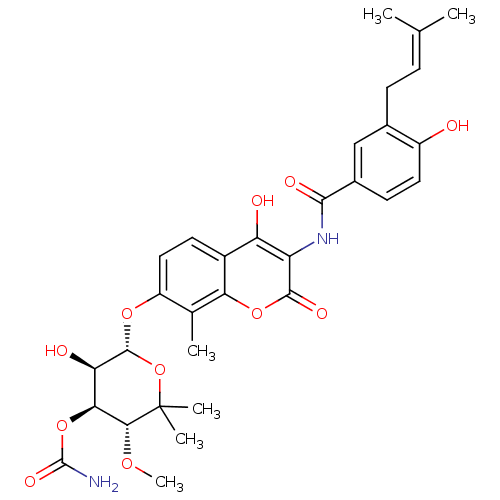
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jakopin, ?; Ila?, J; Baran?oková, M; Brvar, M; Tammela, P; Sollner Dolenc, M; Toma?i?, T; Kikelj, D Discovery of substituted oxadiazoles as a novel scaffold for DNA gyrase inhibitors. Eur J Med Chem130:171-184 (2017) [PubMed] Article
Jakopin, ?; Ila?, J; Baran?oková, M; Brvar, M; Tammela, P; Sollner Dolenc, M; Toma?i?, T; Kikelj, D Discovery of substituted oxadiazoles as a novel scaffold for DNA gyrase inhibitors. Eur J Med Chem130:171-184 (2017) [PubMed] Article