| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Gamma-aminobutyric acid receptor subunit alpha-3 |
|---|
| Ligand | BDBM50000687 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 34.67±n/a nM |
|---|
| Comments | PDSP_1758 |
|---|
| Citation |  Ebert, B; Thompson, SA; Saounatsou, K; McKernan, R; Krogsgaard-Larsen, P; Wafford, KA Differences in agonist/antagonist binding affinity and receptor transduction using recombinant human gamma-aminobutyric acid type A receptors. Mol Pharmacol52:1150-6 (1997) [PubMed] Ebert, B; Thompson, SA; Saounatsou, K; McKernan, R; Krogsgaard-Larsen, P; Wafford, KA Differences in agonist/antagonist binding affinity and receptor transduction using recombinant human gamma-aminobutyric acid type A receptors. Mol Pharmacol52:1150-6 (1997) [PubMed] |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Gamma-aminobutyric acid receptor subunit alpha-3 |
|---|
| Name: | Gamma-aminobutyric acid receptor subunit alpha-3 |
|---|
| Synonyms: | GABA A Alpha3Beta3Gamma2 | GABA A receptor alpha-3/beta-2/gamma-2 | GABA C | GABA receptor alpha-3 subunit | GABA-A receptor | GABRA3 | GBRA3_HUMAN | Gamma-aminobutyric acid receptor subunit alpha-3 | agonist GABA site |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 55175.47 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | GABA A Alpha3Beta1Gamma2 0 HUMAN::P34903 |
|---|
| Residue: | 492 |
|---|
| Sequence: | MIITQTSHCYMTSLGILFLINILPGTTGQGESRRQEPGDFVKQDIGGLSPKHAPDIPDDS
TDNITIFTRILDRLLDGYDNRLRPGLGDAVTEVKTDIYVTSFGPVSDTDMEYTIDVFFRQ
TWHDERLKFDGPMKILPLNNLLASKIWTPDTFFHNGKKSVAHNMTTPNKLLRLVDNGTLL
YTMRLTIHAECPMHLEDFPMDVHACPLKFGSYAYTTAEVVYSWTLGKNKSVEVAQDGSRL
NQYDLLGHVVGTEIIRSSTGEYVVMTTHFHLKRKIGYFVIQTYLPCIMTVILSQVSFWLN
RESVPARTVFGVTTVLTMTTLSISARNSLPKVAYATAMDWFIAVCYAFVFSALIEFATVN
YFTKRSWAWEGKKVPEALEMKKKTPAAPAKKTSTTFNIVGTTYPINLAKDTEFSTISKGA
APSASSTPTIIASPKATYVQDSPTETKTYNSVSKVDKISRIIFPVLFAIFNLVYWATYVN
RESAIKGMIRKQ
|
|
|
|---|
| BDBM50000687 |
|---|
| n/a |
|---|
| Name | BDBM50000687 |
|---|
| Synonyms: | 2-(3-Carboxy-propyl)-6-(4-methoxy-phenyl)-2H-pyridazin-3-ylidene-ammonium; bromide(gabazine) | 4-[6-Amino-3-(4-methoxy-phenyl)-6H-pyridazin-1-yl]-butyric acid : bromide | 6-Amino-1-(3-carboxy-propyl)-3-(4-methoxy-phenyl)-pyridazin-1-ium; bromide | CHEMBL303580 | SR 95531 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H18N3O3 |
|---|
| Mol. Mass. | 288.3212 |
|---|
| SMILES | COc1ccc(cc1)-c1ccc(=[NH2+])n(CCCC(O)=O)n1 |
|---|
| Structure | 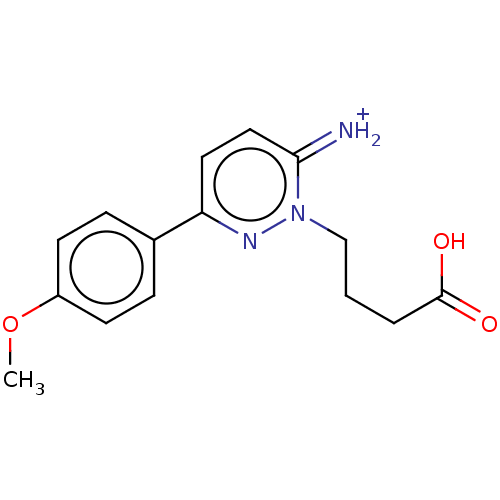
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ebert, B; Thompson, SA; Saounatsou, K; McKernan, R; Krogsgaard-Larsen, P; Wafford, KA Differences in agonist/antagonist binding affinity and receptor transduction using recombinant human gamma-aminobutyric acid type A receptors. Mol Pharmacol52:1150-6 (1997) [PubMed]
Ebert, B; Thompson, SA; Saounatsou, K; McKernan, R; Krogsgaard-Larsen, P; Wafford, KA Differences in agonist/antagonist binding affinity and receptor transduction using recombinant human gamma-aminobutyric acid type A receptors. Mol Pharmacol52:1150-6 (1997) [PubMed]