| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Ligand | BDBM50094703 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | >10000±n/a nM |
|---|
| Comments | PDSP_2986 |
|---|
| Citation |  Jarvis, MF; Yu, H; Kohlhaas, K; Alexander, K; Lee, CH; Jiang, M; Bhagwat, SS; Williams, M; Kowaluk, EA ABT-702 (4-amino-5-(3-bromophenyl)-7-(6-morpholinopyridin-3-yl)pyrido[2, 3-d]pyrimidine), a novel orally effective adenosine kinase inhibitor with analgesic and anti-inflammatory properties: I. In vitro characterization and acute antinociceptive effects in the mouse. J Pharmacol Exp Ther295:1156-64 (2000) [PubMed] Jarvis, MF; Yu, H; Kohlhaas, K; Alexander, K; Lee, CH; Jiang, M; Bhagwat, SS; Williams, M; Kowaluk, EA ABT-702 (4-amino-5-(3-bromophenyl)-7-(6-morpholinopyridin-3-yl)pyrido[2, 3-d]pyrimidine), a novel orally effective adenosine kinase inhibitor with analgesic and anti-inflammatory properties: I. In vitro characterization and acute antinociceptive effects in the mouse. J Pharmacol Exp Ther295:1156-64 (2000) [PubMed] |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Name: | Voltage-dependent L-type calcium channel subunit alpha-1C |
|---|
| Synonyms: | CAC1C_RAT | Cach2 | Cacn2 | Cacna1c | Cacnl1a1 | Calcium channel | Calcium channel (Type L) | Calcium channel diltiazem | Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle | Cchl1a1 | RBC | Rat brain class C | Voltage-dependent L-type calcium channel subunit alpha-1C | Voltage-gated L-type calcium channel | Voltage-gated L-type calcium channel alpha-1C subunit | Voltage-gated calcium channel subunit alpha Cav1.2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 243492.26 |
|---|
| Organism: | RAT |
|---|
| Description: | Calcium channel 0 RAT::P22002 |
|---|
| Residue: | 2169 |
|---|
| Sequence: | MIRAFAQPSTPPYQPLSSCLSEDTERKFKGKVVHEAQLNCFYISPGGSNYGSPRPAHANM
NANAAAGLAPEHIPTPGAALSWLAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK
KQGGTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPED
DSNATNSNLERVEYLFLIIFTVEAFLKVIAYGLLFHPNAYLRNGWNLLDFIIVVVGLFSA
ILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH
IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIIDVPAEEDPSPCALETGHGRQCQNGT
VCKPGWDGPKHGITNFDNFAFAMLTVFQCITMEGWTDVLYWMQDAMGYELPWVYFVSLVI
FGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE
NEDEGMDEDKPRNMSMPTSETESVNTENVAGGDIEGENCGARLAHRISKSKFSRYWRRWN
RFCRRKCRAAVKSNVFYWLVIFLVFLNTLTIASEHYNQPHWLTEVQDTANKALLALFTAE
MLLKMYSLGLQAYFVSLFNRFDCFIVCGGILETILVETKIMSPLGISCWRCVRLLRIFKI
TRYWNSLSNLVASLLNSLRSIASLLLLLFLFIIIFSLLGMQLFGGKFNFDEMQTRRSTFD
NFPQSLLTVFQILTGEDWNSVMYDGIMAYGGPSFPGMLVCIYFIILFISPNYILLNLFLA
IAVDNLADAESLTSAQKEEEEEKERKKLARTASPEKKQEVMEKPAVEESKEEKIELKSIT
ADGESPPTTKINMDDLQPSENEDKSPHSNPDTAGEEDEEEPEMPVGPRPRPLSELHLKEK
AVPMPEASAFFIFSPNNRFRLQCHRIVNDTIFTNLILFFILLSSISLAAEDPVQHTSFRN
HILFYFDIVFTTIFTIEIALKMTAYGAFLHKGSFCRNYFNILDLLVVSVSLISFGIQSSA
INVVKILRVLRVLRPLRINRAKGLKHVVQCVFVAIRTIGNIVIVTTLLQFMFACIGVQLF
KGKLYTCSDSSKQTEAESKGNYITYKTGEVDHPIIQPRSWENSKFDFDNVLAAMMALFTV
STFEGWPELLYRSIDSHTEDKGPIYNYRVEISIFFIIYIIIIAFFMMNIFVGFVIVTFQE
QGEQEYKNCELDKNQRQCVEYALKARPLPRYIPKNQHQYKVWYVVNSTYFEYLMFVLILL
NTICLAMQHYGQSCLFKIAMNILNMLFTGLFTVEMILKLIAFKPKHYFCDAWNTFDALIV
VGSIVDIAITEVHPAEHTQCSPSMSAEENSRISITFFRLFRVMRLVKLLSRGEGIRTLLW
TFIKSFQALPYVALLIVMLFFIYAVIGMQVFGKIALNDTTEINRNNNFQTFPQAVLLLFR
CATGEAWQDIMLACMPGKKCAPESEPSNSTEGETPCGSSFAVFYFISFYMLCAFLIINLF
VAVIMDNFDYLTRDWSILGPHHLDEFKRIWAEYDPEAKGRIKHLDVVTLLRRIQPPLGFG
KLCPHRVACKRLVSMNMPLNSDGTVMFNATLFALVRTALRIKTEGNLEQANEELRAIIKK
IWKRTSMKLLDQVVPPAGDDEVTVGKFYATFLIQEYFRKFKKRKEQGLVGKPSQRNALSL
QAGLRTLHDIGPEIRRAISGDLTAEEELDKAMKEAVSAASEDDIFRRAGGLFGNHVSYYQ
SDSRSNFPQTFATQRPLHINKTGNNQADTESPSHEKLVDSTFTPSSYSSTGSNANINNAN
NTALGRFPHPAGYSSTVSTVEGHGPPLSPAVRVQEAAWKLSSKRCHSRESQGATVSQDMF
PDETRSSVRLSEEVEYCSEPSLLSTDILSYQDDENRQLTCLEEDKREIQPCPKRSFLRSA
SLGRRASFHLECLKRQKDQGGDISQKTALPLHLVHHQALAVAGLSPLLQRSHSPSTFPRP
RPTPPVTPGSRGRPLQPIPTLRLEGAESSEKLNSSFPSIHCSSWSEETTACSGGSSMARR
ARPVSLTVPSQAGAPGRQFHGSASSLVEAVLISEGLGQFAQDPKFIEVTTQELADACDMT
IEEMENAADNILSGGAQQSPNGTLLPFVNCRDPGQDRAVVPEDESCVYALGRGRSEEALP
DSRSYVSNL
|
|
|
|---|
| BDBM50094703 |
|---|
| n/a |
|---|
| Name | BDBM50094703 |
|---|
| Synonyms: | 5-(3-Bromo-phenyl)-7-(6-morpholin-4-yl-pyridin-3-yl)-pyrido[2,3-d]pyrimidin-4-ylamine | 5-(3-bromophenyl)-7-(6-morpholin-4-ylpyridin-3-yl)pyrido[2,3-d]pyrimidin-4-ylamine | ABT-702 | CHEMBL66089 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H19BrN6O |
|---|
| Mol. Mass. | 463.33 |
|---|
| SMILES | Nc1ncnc2nc(cc(-c3cccc(Br)c3)c12)-c1ccc(nc1)N1CCOCC1 |
|---|
| Structure | 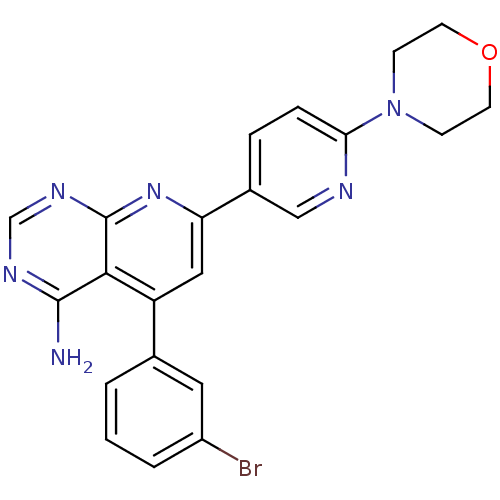
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jarvis, MF; Yu, H; Kohlhaas, K; Alexander, K; Lee, CH; Jiang, M; Bhagwat, SS; Williams, M; Kowaluk, EA ABT-702 (4-amino-5-(3-bromophenyl)-7-(6-morpholinopyridin-3-yl)pyrido[2, 3-d]pyrimidine), a novel orally effective adenosine kinase inhibitor with analgesic and anti-inflammatory properties: I. In vitro characterization and acute antinociceptive effects in the mouse. J Pharmacol Exp Ther295:1156-64 (2000) [PubMed]
Jarvis, MF; Yu, H; Kohlhaas, K; Alexander, K; Lee, CH; Jiang, M; Bhagwat, SS; Williams, M; Kowaluk, EA ABT-702 (4-amino-5-(3-bromophenyl)-7-(6-morpholinopyridin-3-yl)pyrido[2, 3-d]pyrimidine), a novel orally effective adenosine kinase inhibitor with analgesic and anti-inflammatory properties: I. In vitro characterization and acute antinociceptive effects in the mouse. J Pharmacol Exp Ther295:1156-64 (2000) [PubMed]