| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Muscarinic acetylcholine receptor M3 |
|---|
| Ligand | BDBM50048803 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | >10000±n/a nM |
|---|
| Comments | PDSP_1996 |
|---|
| Citation |  Kroeze, WK; Hufeisen, SJ; Popadak, BA; Renock, SM; Steinberg, S; Ernsberger, P; Jayathilake, K; Meltzer, HY; Roth, BL H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology28:519-26 (2003) [PubMed] Article Kroeze, WK; Hufeisen, SJ; Popadak, BA; Renock, SM; Steinberg, S; Ernsberger, P; Jayathilake, K; Meltzer, HY; Roth, BL H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology28:519-26 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Muscarinic acetylcholine receptor M3 |
|---|
| Name: | Muscarinic acetylcholine receptor M3 |
|---|
| Synonyms: | ACM3_HUMAN | CHRM3 | Cholinergic, muscarinic M3 | Muscarinic Receptors M3 | Muscarinic receptor M3 | RecName: Full=Muscarinic acetylcholine receptor M3 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 66151.03 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P20309 |
|---|
| Residue: | 590 |
|---|
| Sequence: | MTLHNNSTTSPLFPNISSSWIHSPSDAGLPPGTVTHFGSYNVSRAAGNFSSPDGTTDDPL
GGHTVWQVVFIAFLTGILALVTIIGNILVIVSFKVNKQLKTVNNYFLLSLACADLIIGVI
SMNLFTTYIIMNRWALGNLACDLWLAIDYVASNASVMNLLVISFDRYFSITRPLTYRAKR
TTKRAGVMIGLAWVISFVLWAPAILFWQYFVGKRTVPPGECFIQFLSEPTITFGTAIAAF
YMPVTIMTILYWRIYKETEKRTKELAGLQASGTEAETENFVHPTGSSRSCSSYELQQQSM
KRSNRRKYGRCHFWFTTKSWKPSSEQMDQDHSSSDSWNNNDAAASLENSASSDEEDIGSE
TRAIYSIVLKLPGHSTILNSTKLPSSDNLQVPEEELGMVDLERKADKLQAQKSVDDGGSF
PKSFSKLPIQLESAVDTAKTSDVNSSVGKSTATLPLSFKEATLAKRFALKTRSQITKRKR
MSLVKEKKAAQTLSAILLAFIITWTPYNIMVLVNTFCDSCIPKTFWNLGYWLCYINSTVN
PVCYALCNKTFRTTFKMLLLCQCDKKKRRKQQYQQRQSVIFHKRAPEQAL
|
|
|
|---|
| BDBM50048803 |
|---|
| n/a |
|---|
| Name | BDBM50048803 |
|---|
| Synonyms: | 5-(2-(4-(benzo[d]isothiazol-3-yl)piperazin-1-yl)ethyl)-6-chloroindolin-2-one | 5-[2-(4-Benzo[d]isothiazol-3-yl-piperazin-1-yl)-ethyl]-6-chloro-1,3-dihydro-indol-2-one | 5-[2-(4-Benzo[d]isothiazol-3-yl-piperazin-1-yl)-ethyl]-6-chloro-1,3-dihydro-indol-2-one (Ziprasidone) | 5-[2-(4-Benzo[d]isothiazol-3-yl-piperazin-1-yl)-ethyl]-6-chloro-1,3-dihydro-indol-2-one(Norastemizole) | CHEMBL708 | GEODON | ZIPRASIDONE | ZIPRASIDONE HYDROCHLORIDE |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H21ClN4OS |
|---|
| Mol. Mass. | 412.936 |
|---|
| SMILES | Clc1cc2NC(=O)Cc2cc1CCN1CCN(CC1)c1nsc2ccccc12 |
|---|
| Structure | 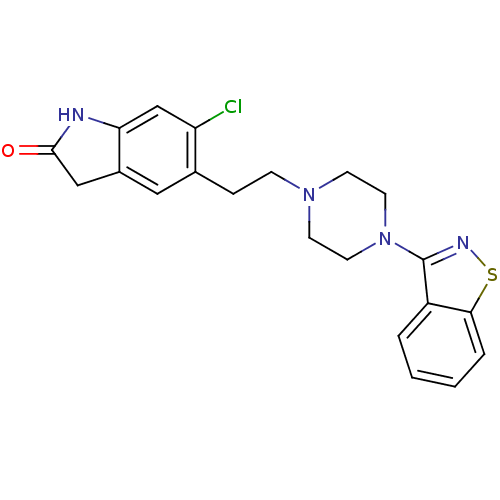
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kroeze, WK; Hufeisen, SJ; Popadak, BA; Renock, SM; Steinberg, S; Ernsberger, P; Jayathilake, K; Meltzer, HY; Roth, BL H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology28:519-26 (2003) [PubMed] Article
Kroeze, WK; Hufeisen, SJ; Popadak, BA; Renock, SM; Steinberg, S; Ernsberger, P; Jayathilake, K; Meltzer, HY; Roth, BL H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology28:519-26 (2003) [PubMed] Article