

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | ||
| Ligand | BDBM14773 | ||
| Substrate/Competitor | n/a | ||
| Ki | 200±n/a nM | ||
| Comments | PDSP_3036 | ||
| Citation |  Zhao, Y; Zhang, HT; O'Donnell, JM Inhibitor binding to type 4 phosphodiesterase (PDE4) assessed using [3H]piclamilast and [3H]rolipram. J Pharmacol Exp Ther305:565-72 (2003) [PubMed] Article Zhao, Y; Zhang, HT; O'Donnell, JM Inhibitor binding to type 4 phosphodiesterase (PDE4) assessed using [3H]piclamilast and [3H]rolipram. J Pharmacol Exp Ther305:565-72 (2003) [PubMed] Article | ||
| More Info.: | Get all data from this article | ||
| cAMP-specific 3',5'-cyclic phosphodiesterase 4B | |||
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4B | ||
| Synonyms: | DPDE4 | PDE4B_RAT | Pde4b | Phosphodiesterase 4B | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 83349.50 | ||
| Organism: | RAT | ||
| Description: | P14646 | ||
| Residue: | 736 | ||
| Sequence: |
| ||
| BDBM14773 | |||
| n/a | |||
| Name | BDBM14773 | ||
| Synonyms: | 4-cyano-4-(3-cyclopentyloxy-4-methoxy-phenyl)cyclohexane-1-carboxylic acid | 4-cyano-4-[3-(cyclopentyloxy)-4-methoxyphenyl]cyclohexane-1-carboxylic acid | Ariflo | Cilomilast | SB207499 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H25NO4 | ||
| Mol. Mass. | 343.4168 | ||
| SMILES | COc1ccc(cc1OC1CCCC1)C1(CCC(CC1)C(O)=O)C#N |(1.4,-2.1,;1.4,-.56,;.07,.21,;-1.27,-.56,;-2.6,.21,;-2.6,1.75,;-1.27,2.52,;.07,1.75,;1.4,2.52,;1.4,4.06,;2.65,4.96,;2.17,6.43,;.63,6.43,;.15,4.96,;-3.93,2.52,;-4.44,3.97,;-5.95,4.26,;-6.96,3.1,;-6.46,1.65,;-4.94,1.35,;-8.47,3.39,;-9.48,2.23,;-8.97,4.85,;-3.16,3.85,;-2.39,5.18,)| | ||
| Structure | 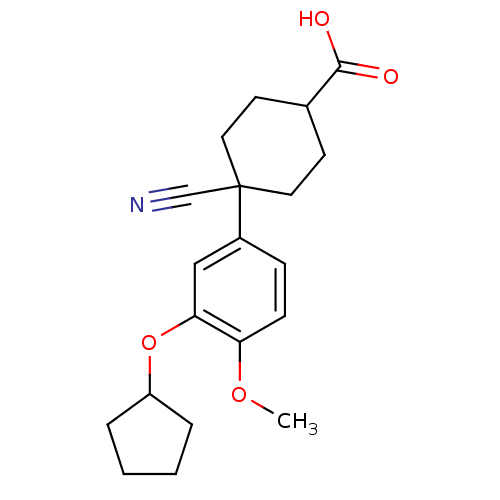
| ||