| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Promotilin |
|---|
| Ligand | BDBM86675 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | 50±n/a nM |
|---|
| Comments | PDSP_6066 |
|---|
| Citation |  Matsuura, B; Dong, M; Coulie, B; Pinon, DI; Miller, LJ Demonstration of a specific site of covalent labeling of the human motilin receptor using a biologically active photolabile motilin analog. J Pharmacol Exp Ther313:1101-8 (2005) [PubMed] Article Matsuura, B; Dong, M; Coulie, B; Pinon, DI; Miller, LJ Demonstration of a specific site of covalent labeling of the human motilin receptor using a biologically active photolabile motilin analog. J Pharmacol Exp Ther313:1101-8 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Promotilin |
|---|
| Name: | Promotilin |
|---|
| Synonyms: | MLN | MOTI_HUMAN | Motilin | Promotilin |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 12919.83 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Motilin 0 HUMAN::P12872 |
|---|
| Residue: | 115 |
|---|
| Sequence: | MVSRKAVAALLVVHVAAMLASQTEAFVPIFTYGELQRMQEKERNKGQKKSLSVWQRSGEE
GPVDPAEPIREEENEMIKLTAPLEIGMRMNSRQLEKYPATLEGLLSEMLPQHAAK
|
|
|
|---|
| BDBM86675 |
|---|
| n/a |
|---|
| Name | BDBM86675 |
|---|
| Synonyms: | CAS_0 | NSC_0 | [Bpa5,Ile13]Motilin | [L-Bpa5,Ile13]Motilin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C129H196N34O35 |
|---|
| Mol. Mass. | 2783.1433 |
|---|
| SMILES | [#6]-[#6]-[#6](-[#6])-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#7])=O)-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6]-[#6](-[#8])=O)-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccc(-[#8])cc1)-[#7]-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccc(cc1)-[#6](=O)-c1ccccc1)-[#7]-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@@H]-1-[#6]-[#6]-[#6]-[#7]-1-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@@H](-[#7])-[#6]-c1ccccc1)-[#6](-[#6])-[#6])-[#6@@H](-[#6])-[#6]-[#6])-[#6@@H](-[#6])-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#6](-[#6])=[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#8])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](/[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6](-[#7])=O)-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6](-[#7])=O)-[#6](-[#8])=O |r,w:127.132| |
|---|
| Structure | 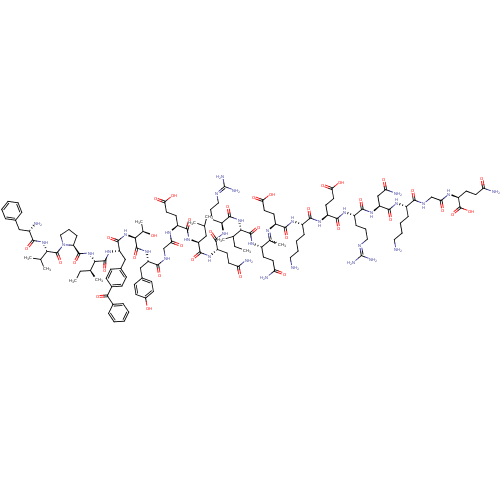
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Matsuura, B; Dong, M; Coulie, B; Pinon, DI; Miller, LJ Demonstration of a specific site of covalent labeling of the human motilin receptor using a biologically active photolabile motilin analog. J Pharmacol Exp Ther313:1101-8 (2005) [PubMed] Article
Matsuura, B; Dong, M; Coulie, B; Pinon, DI; Miller, LJ Demonstration of a specific site of covalent labeling of the human motilin receptor using a biologically active photolabile motilin analog. J Pharmacol Exp Ther313:1101-8 (2005) [PubMed] Article