| Reaction Details |
|---|
 | Report a problem with these data |
| Target | V-type proton ATPase subunit c' |
|---|
| Ligand | BDBM53623 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Cherry Pick 2 |
|---|
| EC50 | 1404±n/a nM |
|---|
| Citation |  PubChem, PC Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Cherry Pick 2 PubChem Bioassay(2012)[AID] PubChem, PC Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Cherry Pick 2 PubChem Bioassay(2012)[AID] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| V-type proton ATPase subunit c' |
|---|
| Name: | V-type proton ATPase subunit c' |
|---|
| Synonyms: | CLS9 | TFP3 | VATL2_YEAST | VMA11 | Vma11p |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 17039.28 |
|---|
| Organism: | Saccharomyces cerevisiae S288c |
|---|
| Description: | gi_6325022 |
|---|
| Residue: | 164 |
|---|
| Sequence: | MSTQLASNIYAPLYAPFFGFAGCAAAMVLSCLGAAIGTAKSGIGIAGIGTFKPELIMKSL
IPVVMSGILAIYGLVVAVLIAGNLSPTEDYTLFNGFMHLSCGLCVGFACLSSGYAIGMVG
DVGVRKYMHQPRLFVGIVLILIFSEVLGLYGMIVALILNTRGSE
|
|
|
|---|
| BDBM53623 |
|---|
| n/a |
|---|
| Name | BDBM53623 |
|---|
| Synonyms: | 5-chloranyl-2-(4-chlorophenyl)-4-methyl-1,2-thiazol-3-one | 5-chloro-2-(4-chlorophenyl)-4-methyl-1,2-thiazol-3-one | 5-chloro-2-(4-chlorophenyl)-4-methyl-3(2H)-isothiazolone | 5-chloro-2-(4-chlorophenyl)-4-methyl-3-isothiazolone | 5-chloro-2-(4-chlorophenyl)-4-methyl-4-isothiazolin-3-one | MLS000684518 | SMR000269693 | cid_759209 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H7Cl2NOS |
|---|
| Mol. Mass. | 260.14 |
|---|
| SMILES | Cc1c(Cl)sn(-c2ccc(Cl)cc2)c1=O |
|---|
| Structure | 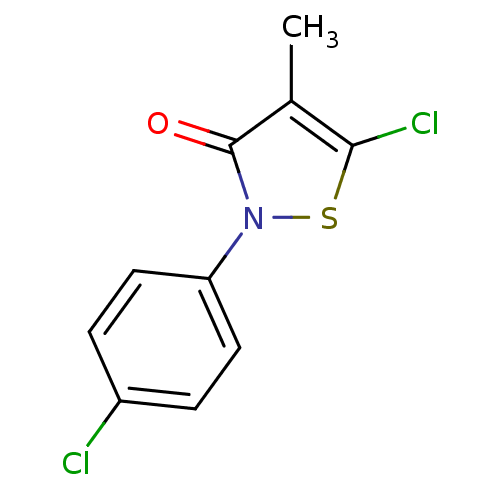
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Cherry Pick 2 PubChem Bioassay(2012)[AID]
PubChem, PC Dose Response of Small Molecules that Regulate V-ATPase Proton Transport in Yeast using pHLuorin, Cherry Pick 2 PubChem Bioassay(2012)[AID]