| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA dC->dU-editing enzyme APOBEC-3A |
|---|
| Ligand | BDBM41028 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | SAR analysis of small molecule inhibitors of APOBEC3A DNA Deaminase via a fluorescence-based single-stranded DNA deaminase assay - Set 2 |
|---|
| Temperature | 298.15±n/a K |
|---|
| IC50 | 1210±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  PubChem, PC SAR analysis of small molecule inhibitors of APOBEC3A DNA Deaminase via a fluorescence-based single-stranded DNA deaminase assay - Set 2 PubChem Bioassay(2012)[AID] PubChem, PC SAR analysis of small molecule inhibitors of APOBEC3A DNA Deaminase via a fluorescence-based single-stranded DNA deaminase assay - Set 2 PubChem Bioassay(2012)[AID] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA dC->dU-editing enzyme APOBEC-3A |
|---|
| Name: | DNA dC->dU-editing enzyme APOBEC-3A |
|---|
| Synonyms: | ABC3A_HUMAN | APOBEC3A | probable DNA dC->dU-editing enzyme APOBEC-3A |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 23013.77 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_21955158 |
|---|
| Residue: | 199 |
|---|
| Sequence: | MEASPASGPRHLMDPHIFTSNFNNGIGRHKTYLCYEVERLDNGTSVKMDQHRGFLHNQAK
NLLCGFYGRHAELRFLDLVPSLQLDPAQIYRVTWFISWSPCFSWGCAGEVRAFLQENTHV
RLRIFAARIYDYDPLYKEALQMLRDAGAQVSIMTYDEFKHCWDTFVDHQGCPFQPWDGLD
EHSQALSGRLRAILQNQGN
|
|
|
|---|
| BDBM41028 |
|---|
| n/a |
|---|
| Name | BDBM41028 |
|---|
| Synonyms: | 2-[(5Z)-5-[(E)-3-(2-furanyl)prop-2-enylidene]-4-oxo-2-sulfanylidene-3-thiazolidinyl]propanoic acid | 2-[(5Z)-5-[(E)-3-(2-furyl)prop-2-enylidene]-4-keto-2-thioxo-thiazolidin-3-yl]propionic acid | 2-[(5Z)-5-[(E)-3-(furan-2-yl)prop-2-enylidene]-4-oxidanylidene-2-sulfanylidene-1,3-thiazolidin-3-yl]propanoic acid | 2-[(5Z)-5-[(E)-3-(furan-2-yl)prop-2-enylidene]-4-oxo-2-sulfanylidene-1,3-thiazolidin-3-yl]propanoic acid | 2-{5-[(E)-3-Furan-2-yl-prop-2-en-(Z)-ylidene]-4-oxo-2-thioxo-thiazolidin-3-yl}-propionic acid | MLS000551724 | SMR000145649 | cid_6375556 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H11NO4S2 |
|---|
| Mol. Mass. | 309.361 |
|---|
| SMILES | CC(N1C(=S)S\C(=C/C=C/c2ccco2)C1=O)C(O)=O |
|---|
| Structure | 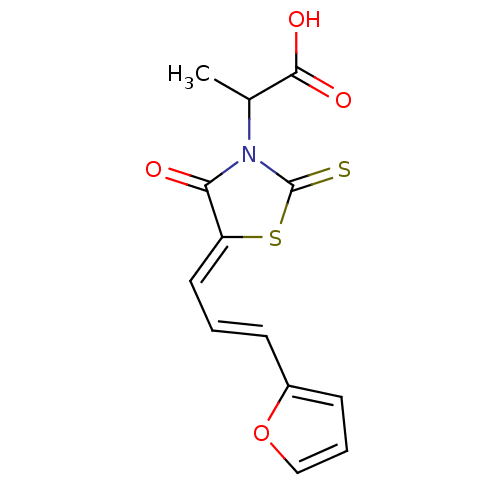
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC SAR analysis of small molecule inhibitors of APOBEC3A DNA Deaminase via a fluorescence-based single-stranded DNA deaminase assay - Set 2 PubChem Bioassay(2012)[AID]
PubChem, PC SAR analysis of small molecule inhibitors of APOBEC3A DNA Deaminase via a fluorescence-based single-stranded DNA deaminase assay - Set 2 PubChem Bioassay(2012)[AID]