| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 130 |
|---|
| Ligand | BDBM92569 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Binding Assay |
|---|
| Kd | 4.9e+3± 3e+2 nM |
|---|
| Citation |  Podust, LM; Ouellet, H; von Kries, JP; de Montellano, PR Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines. J Biol Chem284:25211-9 (2009) [PubMed] Article Podust, LM; Ouellet, H; von Kries, JP; de Montellano, PR Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines. J Biol Chem284:25211-9 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 130 |
|---|
| Name: | Cytochrome P450 130 |
|---|
| Synonyms: | CP130_MYCTU | Cytochrome P450 130 (G243A) | Putative cytochrome P450 130 | cyp130 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 44574.98 |
|---|
| Organism: | Mycobacterium tuberculosis |
|---|
| Description: | P9WPN5 |
|---|
| Residue: | 405 |
|---|
| Sequence: | MTSVMSHEFQLATAETWPNPWPMYRALRDHDPVHHVVPPQRPEYDYYVLSRHADVWSAAR
DHQTFSSAQGLTVNYGELEMIGLHDTPPMVMQDPPVHTEFRKLVSRGFTPRQVETVEPTV
RKFVVERLEKLRANGGGDIVTELFKPLPSMVVAHYLGVPEEDWTQFDGWTQAIVAANAVD
GATTGALDAVGSMMAYFTGLIERRRTEPADDAISHLVAAGVGADGDTAGTLSILAFTFTM
VTGGNDTVTGMLGGSMPLLHRRPDQRRLLLDDPEGIPDAVEELLRLTSPVQGLARTTTRD
VTIGDTTIPAGRRVLLLYGSANRDERQYGPDAAELDVTRCPRNILTFSHGAHHCLGAAAA
RMQCRVALTELLARCPDFEVAESRIVWSGGSYVRRPLSVPFRVTS
|
|
|
|---|
| BDBM92569 |
|---|
| n/a |
|---|
| Name | BDBM92569 |
|---|
| Synonyms: | Arylamine compound 2 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H15N3O2S |
|---|
| Mol. Mass. | 361.417 |
|---|
| SMILES | Nc1ccc(Sc2ccc(cc2)N2C(=O)c3ccc(N)cc3C2=O)cc1 |
|---|
| Structure | 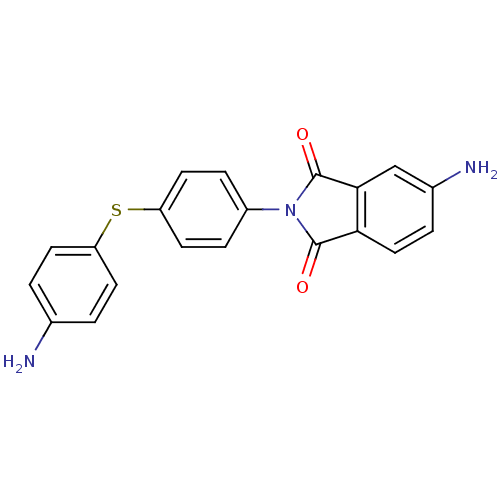
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Podust, LM; Ouellet, H; von Kries, JP; de Montellano, PR Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines. J Biol Chem284:25211-9 (2009) [PubMed] Article
Podust, LM; Ouellet, H; von Kries, JP; de Montellano, PR Interaction of Mycobacterium tuberculosis CYP130 with heterocyclic arylamines. J Biol Chem284:25211-9 (2009) [PubMed] Article