| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Probable nicotinate-nucleotide adenylyltransferase |
|---|
| Ligand | BDBM98114 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | NaMNAT HPLC Enzyme Assay |
|---|
| pH | 7.5±0 |
|---|
| Temperature | 273.15±0 K |
|---|
| IC50 | 20400±0.0 nM |
|---|
| Citation |  Moro, WB; Yang, Z; Kane, TA; Zhou, Q; Harville, S; Brouillette, CG; Brouillette, WJ SAR studies for a new class of antibacterial NAD biosynthesis inhibitors. J Comb Chem11:617-25 (2009) [PubMed] Article Moro, WB; Yang, Z; Kane, TA; Zhou, Q; Harville, S; Brouillette, CG; Brouillette, WJ SAR studies for a new class of antibacterial NAD biosynthesis inhibitors. J Comb Chem11:617-25 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Probable nicotinate-nucleotide adenylyltransferase |
|---|
| Name: | Probable nicotinate-nucleotide adenylyltransferase |
|---|
| Synonyms: | BAMEG_4595 | Deamido-NAD(+) diphosphorylase | Deamido-NAD(+) pyrophosphorylase | NADD_BACAC | Nicotinate mononucleotide adenylyltransferase | Nicotinate-nucleotide adenylyltransferase | Nicotinate-nucleotide adenylyltransferase (NadD) | Nicotinic acid mononucleotide adenylyltransferase (NaMNAT) | nadD |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 21955.08 |
|---|
| Organism: | Bacillus anthracis |
|---|
| Description: | C3L5T6 |
|---|
| Residue: | 189 |
|---|
| Sequence: | MRKIGIIGGTFDPPHYGHLLIANEVYHALNLEEVWFLPNQIPPHKQGRNITSVESRLQML
ELATEAEEHFSICLEELSRKGPSYTYDTMLQLTKKYPDVQFHFIIGGDMVEYLPKWYNIE
ALLDLVTFVGVARPGYKLRTPYPITTVEIPEFAVSSSLLRERYKEKKTCKYLLPEKVQVY
IERNGLYES
|
|
|
|---|
| BDBM98114 |
|---|
| n/a |
|---|
| Name | BDBM98114 |
|---|
| Synonyms: | 1-[4-[(2-chlorophenyl)sulfonylamino]phenyl]-3-(3-nitrophenyl)urea | Compound ID 7{2,8} |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H15ClN4O5S |
|---|
| Mol. Mass. | 446.864 |
|---|
| SMILES | [O-][N+](=O)c1cccc(NC(=O)Nc2ccc(NS(=O)(=O)c3ccccc3Cl)cc2)c1 |
|---|
| Structure | 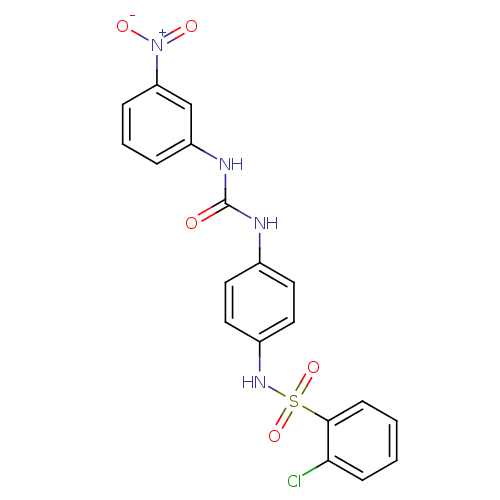
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Moro, WB; Yang, Z; Kane, TA; Zhou, Q; Harville, S; Brouillette, CG; Brouillette, WJ SAR studies for a new class of antibacterial NAD biosynthesis inhibitors. J Comb Chem11:617-25 (2009) [PubMed] Article
Moro, WB; Yang, Z; Kane, TA; Zhou, Q; Harville, S; Brouillette, CG; Brouillette, WJ SAR studies for a new class of antibacterial NAD biosynthesis inhibitors. J Comb Chem11:617-25 (2009) [PubMed] Article