| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Guanylate cyclase soluble subunit alpha-1 |
|---|
| Ligand | BDBM280969 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | sGC Binding Assay |
|---|
| pH | 7.4±n/a |
|---|
| Ki | 0.063±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Berger, R; Chen, Y; Li, G; Garfunkle, J; Li, H; Miao, S; Raghavan, S; Smith, CJ; Stelmach, J; Whitehead, A; Zhang, R; Zhang, Y; Fu, J; Ji, G; Jiang, F Soluble guanylate cyclase stimulators US Patent US10030027 Publication Date 7/24/2018 Berger, R; Chen, Y; Li, G; Garfunkle, J; Li, H; Miao, S; Raghavan, S; Smith, CJ; Stelmach, J; Whitehead, A; Zhang, R; Zhang, Y; Fu, J; Ji, G; Jiang, F Soluble guanylate cyclase stimulators US Patent US10030027 Publication Date 7/24/2018 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Guanylate cyclase soluble subunit alpha-1 |
|---|
| Name: | Guanylate cyclase soluble subunit alpha-1 |
|---|
| Synonyms: | GCS-alpha-1 | GCS-alpha-3 | GCYA1_HUMAN | GUC1A3 | GUCSA3 | GUCY1A1 | GUCY1A3 | Guanylate cyclase soluble (GCS-alpha-1) | Guanylate cyclase soluble subunit alpha-3 | Solube guanylate cyclase alpha-1 (sGC alpha-1) | Solube guanylate cyclase alpha-1 (sGCalpha) | soluble guanylate cyclase alpha-1 (sGC) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 77457.21 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q02108 |
|---|
| Residue: | 690 |
|---|
| Sequence: | MFCTKLKDLKITGECPFSLLAPGQVPNESSEEAAGSSESCKATVPICQDIPEKNIQESLP
QRKTSRSRVYLHTLAESICKLIFPEFERLNVALQRTLAKHKIKESRKSLEREDFEKTIAE
QAVAAGVPVEVIKESLGEEVFKICYEEDENILGVVGGTLKDFLNSFSTLLKQSSHCQEAG
KRGRLEDASILCLDKEDDFLHVYYFFPKRTTSLILPGIIKAAAHVLYETEVEVSLMPPCF
HNDCSEFVNQPYLLYSVHMKSTKPSLSPSKPQSSLVIPTSLFCKTFPFHFMFDKDMTILQ
FGNGIRRLMNRRDFQGKPNFEEYFEILTPKINQTFSGIMTMLNMQFVVRVRRWDNSVKKS
SRVMDLKGQMIYIVESSAILFLGSPCVDRLEDFTGRGLYLSDIPIHNALRDVVLIGEQAR
AQDGLKKRLGKLKATLEQAHQALEEEKKKTVDLLCSIFPCEVAQQLWQGQVVQAKKFSNV
TMLFSDIVGFTAICSQCSPLQVITMLNALYTRFDQQCGELDVYKVETIGDAYCVAGGLHK
ESDTHAVQIALMALKMMELSDEVMSPHGEPIKMRIGLHSGSVFAGVVGVKMPRYCLFGNN
VTLANKFESCSVPRKINVSPTTYRLLKDCPGFVFTPRSREELPPNFPSEIPGICHFLDAY
QQGTNSKPCFQKKDVEDGNANFLGKASGID
|
|
|
|---|
| BDBM280969 |
|---|
| n/a |
|---|
| Name | BDBM280969 |
|---|
| Synonyms: | 5-{4-[(2H-tetrazol- 5-yl)methyl] phenyl}-4-amino- 2-[6-chloro-2-(2- fluorobenzyl)-1H- indazol-3-yl]-5- methyl-5,7- dihydro-6H- pyrrolo[2,3-d] pyrimidin-6-one | US10030027, Example 244A | US10428076, Example 244A |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H22ClFN10O |
|---|
| Mol. Mass. | 581.003 |
|---|
| SMILES | CC1(C(=O)Nc2nc(nc(N)c12)-c1nn(Cc2ccccc2F)c2cc(Cl)ccc12)c1ccc(Cc2nn[nH]n2)cc1 |
|---|
| Structure | 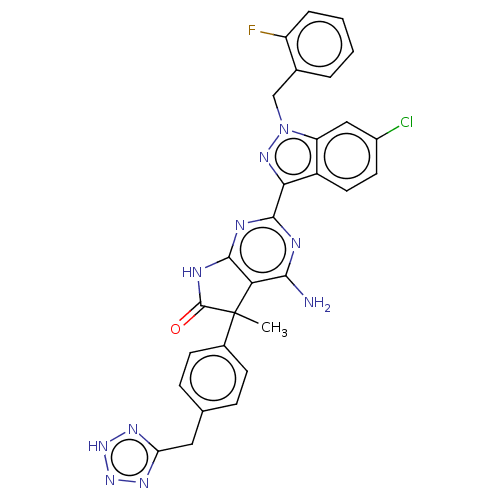
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Berger, R; Chen, Y; Li, G; Garfunkle, J; Li, H; Miao, S; Raghavan, S; Smith, CJ; Stelmach, J; Whitehead, A; Zhang, R; Zhang, Y; Fu, J; Ji, G; Jiang, F Soluble guanylate cyclase stimulators US Patent US10030027 Publication Date 7/24/2018
Berger, R; Chen, Y; Li, G; Garfunkle, J; Li, H; Miao, S; Raghavan, S; Smith, CJ; Stelmach, J; Whitehead, A; Zhang, R; Zhang, Y; Fu, J; Ji, G; Jiang, F Soluble guanylate cyclase stimulators US Patent US10030027 Publication Date 7/24/2018