| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein artemis |
|---|
| Ligand | BDBM66045 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Dose Response confirmation of small molecule inhibitors originally identified via uHTS of Artemis endonuclease activity via a fluorescence intensity assay |
|---|
| IC50 | 11000±n/a nM |
|---|
| Citation |  PubChem, PC Dose Response confirmation of small molecule inhibitors originally identified via uHTS of Artemis endonuclease activity via a fluorescence intensity assay PubChem Bioassay(2013)[AID] PubChem, PC Dose Response confirmation of small molecule inhibitors originally identified via uHTS of Artemis endonuclease activity via a fluorescence intensity assay PubChem Bioassay(2013)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Protein artemis |
|---|
| Name: | Protein artemis |
|---|
| Synonyms: | ARTEMIS | ASCID | DCLRE1C | DCR1C_HUMAN | SCIDA | SNM1C | protein artemis isoform a |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 78427.91 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_76496497 |
|---|
| Residue: | 692 |
|---|
| Sequence: | MSSFEGQMAEYPTISIDRFDRENLRARAYFLSHCHKDHMKGLRAPTLKRRLECSLKVYLY
CSPVTKELLLTSPKYRFWKKRIISIEIETPTQISLVDEASGEKEEIVVTLLPAGHCPGSV
MFLFQGNNGTVLYTGDFRLAQGEAARMELLHSGGRVKDIQSVYLDTTFCDPRFYQIPSRE
ECLSGVLELVRSWITRSPYHVVWLNCKAAYGYEYLFTNLSEELGVQVHVNKLDMFRNMPE
ILHHLTTDRNTQIHACRHPKAEEYFQWSKLPCGITSRNRIPLHIISIKPSTMWFGERSRK
TNVIVRTGESSYRACFSFHSSYSEIKDFLSYLCPVNAYPNVIPVGTTMDKVVEILKPLCR
SSQSTEPKYKPLGKLKRARTVHRDSEEEDDYLFDDPLPIPLRHKVPYPETFHPEVFSMTA
VSEKQPEKLRQTPGCCRAECMQSSRFTNFVDCEESNSESEEEVGIPASLQGDLGSVLHLQ
KADGDVPQWEVFFKRNDEITDESLENFPSSTVAGGSQSPKLFSDSDGESTHISSQNSSQS
THITEQGSQGWDSQSDTVLLSSQERNSGDITSLDKADYRPTIKENIPASLMEQNVICPKD
TYSDLKSRDKDVTIVPSTGEPTTLSSETHIPEEKSLLNLSTNADSQSSSDFEVPSTPEAE
LPKREHLQYLYEKLATGESIAVKKRKCSLLDT
|
|
|
|---|
| BDBM66045 |
|---|
| n/a |
|---|
| Name | BDBM66045 |
|---|
| Synonyms: | (2E)-2-(1-nitrosodecylidene)-1H-pyridine | 1-Pyridin-2-yl-decan-1-one oxime | MLS000555881 | SMR000147398 | cid_5937406 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H24N2O |
|---|
| Mol. Mass. | 248.3639 |
|---|
| SMILES | CCCCCCCCCC(N=O)c1ccccn1 |
|---|
| Structure | 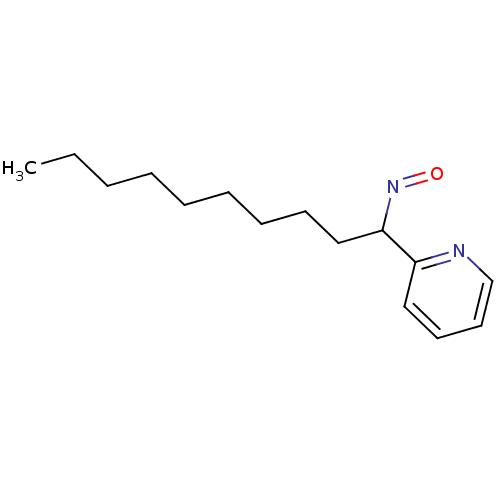
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Dose Response confirmation of small molecule inhibitors originally identified via uHTS of Artemis endonuclease activity via a fluorescence intensity assay PubChem Bioassay(2013)[AID]
PubChem, PC Dose Response confirmation of small molecule inhibitors originally identified via uHTS of Artemis endonuclease activity via a fluorescence intensity assay PubChem Bioassay(2013)[AID]