| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Oxysterols receptor LXR-beta |
|---|
| Ligand | BDBM115003 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Counterscreen for agonists of the daf-12 abnormal Dauer Formation: Luminescence-based cell-based dose response screening assay to identify agonists of the Liver-X-Receptor (LXR) |
|---|
| EC50 | 67568±n/a nM |
|---|
| Citation |  PubChem, PC Counterscreen for agonists of the daf-12 abnormal Dauer Formation: Luminescence-based cell-based dose response screening assay to identify agonists of the Liver-X-Receptor (LXR) PubChem Bioassay(2013)[AID] PubChem, PC Counterscreen for agonists of the daf-12 abnormal Dauer Formation: Luminescence-based cell-based dose response screening assay to identify agonists of the Liver-X-Receptor (LXR) PubChem Bioassay(2013)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Oxysterols receptor LXR-beta |
|---|
| Name: | Oxysterols receptor LXR-beta |
|---|
| Synonyms: | LXRB | Liver X receptor beta (NR1H2) | Liver X, LXR beta | NER | NR1H2 | NR1H2_HUMAN | Nuclear receptor NER | UNR | Ubiquitously-expressed nuclear receptor |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 50978.79 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P55055 |
|---|
| Residue: | 460 |
|---|
| Sequence: | MSSPTTSSLDTPLPGNGPPQPGAPSSSPTVKEEGPEPWPGGPDPDVPGTDEASSACSTDW
VIPDPEEEPERKRKKGPAPKMLGHELCRVCGDKASGFHYNVLSCEGCKGFFRRSVVRGGA
RRYACRGGGTCQMDAFMRRKCQQCRLRKCKEAGMREQCVLSEEQIRKKKIRKQQQESQSQ
SQSPVGPQGSSSSASGPGASPGGSEAGSQGSGEGEGVQLTAAQELMIQQLVAAQLQCNKR
SFSDQPKVTPWPLGADPQSRDARQQRFAHFTELAIISVQEIVDFAKQVPGFLQLGREDQI
ALLKASTIEIMLLETARRYNHETECITFLKDFTYSKDDFHRAGLQVEFINPIFEFSRAMR
RLGLDDAEYALLIAINIFSADRPNVQEPGRVEALQQPYVEALLSYTRIKRPQDQLRFPRM
LMKLVSLRTLSSVHSEQVFALRLQDKKLPPLLSEIWDVHE
|
|
|
|---|
| BDBM115003 |
|---|
| n/a |
|---|
| Name | BDBM115003 |
|---|
| Synonyms: | 2,6-bis(3-methyl-1-pyrazolyl)pyridine | 2,6-bis(3-methyl-1H-pyrazol-1-yl)pyridine | 2,6-bis(3-methylpyrazol-1-yl)pyridine | MLS000702817 | SMR000229839 | cid_850137 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H13N5 |
|---|
| Mol. Mass. | 239.2758 |
|---|
| SMILES | Cc1ccn(n1)-c1cccc(n1)-n1ccc(C)n1 |
|---|
| Structure | 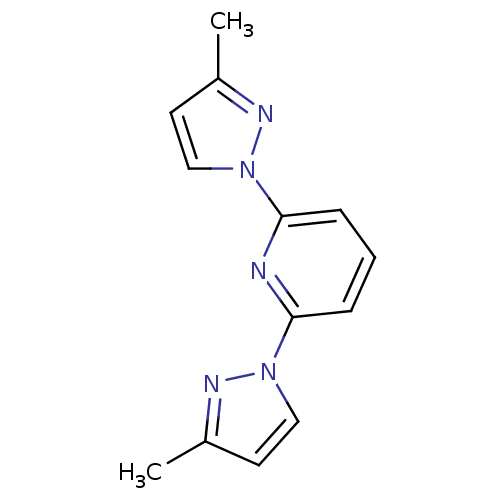
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Counterscreen for agonists of the daf-12 abnormal Dauer Formation: Luminescence-based cell-based dose response screening assay to identify agonists of the Liver-X-Receptor (LXR) PubChem Bioassay(2013)[AID]
PubChem, PC Counterscreen for agonists of the daf-12 abnormal Dauer Formation: Luminescence-based cell-based dose response screening assay to identify agonists of the Liver-X-Receptor (LXR) PubChem Bioassay(2013)[AID]