

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | cGMP-dependent 3',5'-cyclic phosphodiesterase | ||
| Ligand | BDBM131002 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Scintillation Proximity Assay | ||
| IC50 | 1.06±n/a nM | ||
| Citation |  Helal, CJ; Chappie, TA; Humphrey, JM Pyrazolo[3,4-d]pyrimidine compounds and their use as PDE2 inhibitors and/or CYP3A4 inhibitors US Patent US8829010 Publication Date 9/9/2014 Helal, CJ; Chappie, TA; Humphrey, JM Pyrazolo[3,4-d]pyrimidine compounds and their use as PDE2 inhibitors and/or CYP3A4 inhibitors US Patent US8829010 Publication Date 9/9/2014 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| cGMP-dependent 3',5'-cyclic phosphodiesterase | |||
| Name: | cGMP-dependent 3',5'-cyclic phosphodiesterase | ||
| Synonyms: | CGS-PDE | Cyclic GMP-stimulated phosphodiesterase | Homo sapiens phosphodiesterase 2A (PDE2A) | NM_002599 | PDE2A | PDE2A_HUMAN | cGSPDE | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 105691.58 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O00408 | ||
| Residue: | 941 | ||
| Sequence: |
| ||
| BDBM131002 | |||
| n/a | |||
| Name | BDBM131002 | ||
| Synonyms: | US8829010, 53 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H20F3N7 | ||
| Mol. Mass. | 415.4149 | ||
| SMILES | CN(C)c1ncnc2n(C)nc(-c3cnn(C)c3-c3ccc(cc3C)C(F)(F)F)c12 |(-5.29,3.68,;-3.96,2.91,;-3.19,4.25,;-3.96,1.37,;-5.29,.6,;-5.29,-.94,;-3.96,-1.71,;-2.63,-.94,;-1.06,-1.33,;-.5,-2.77,;-.22,-.04,;-1.19,1.16,;-.79,2.64,;-1.27,4.11,;-.02,5.01,;1.23,4.11,;2.69,4.58,;.75,2.64,;1.66,1.4,;1.26,-.09,;2.35,-1.18,;3.83,-.78,;4.23,.71,;3.14,1.8,;3.54,3.28,;4.92,-1.87,;6.41,-1.47,;4.52,-3.36,;6.01,-2.96,;-2.63,.6,)| | ||
| Structure | 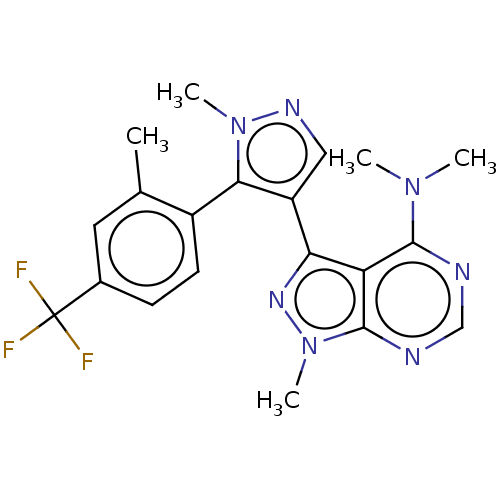
| ||