

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Proto-oncogene tyrosine-protein kinase ROS | ||
| Ligand | BDBM144327 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Inhibition Assay | ||
| IC50 | 0.86±n/a nM | ||
| Citation |  Shimada, I; Kurosawa, K; Matsuya, T; Iikubo, K; Kondoh, Y; Kamikawa, A; Tomiyama, H; Iwai, Y Diamino heterocyclic carboxamide compound US Patent US8969336 Publication Date 3/3/2015 Shimada, I; Kurosawa, K; Matsuya, T; Iikubo, K; Kondoh, Y; Kamikawa, A; Tomiyama, H; Iwai, Y Diamino heterocyclic carboxamide compound US Patent US8969336 Publication Date 3/3/2015 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Proto-oncogene tyrosine-protein kinase ROS | |||
| Name: | Proto-oncogene tyrosine-protein kinase ROS | ||
| Synonyms: | MCF3 | Proto-oncogene c-Ros | Proto-oncogene c-Ros-1 | ROS | ROS1 | ROS1_HUMAN | Receptor tyrosine kinase c-ros oncogene 1 | Tyrosine-Protein Kinase Receptor ROS | c-Ros receptor tyrosine kinase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 263897.39 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P08922 | ||
| Residue: | 2347 | ||
| Sequence: |
| ||
| BDBM144327 | |||
| n/a | |||
| Name | BDBM144327 | ||
| Synonyms: | US8969336, 571 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C31H48N8O2 | ||
| Mol. Mass. | 564.7652 | ||
| SMILES | CCc1nc(C(N)=O)c(Nc2ccc(N3CCC(CC3)N3CCN(C)CC3)c(C)c2)nc1N[C@H]1CC[C@@](C)(O)CC1 |r,wU:36.40,wD:36.41,33.36,(3.3,-7.91,;3.3,-6.37,;1.97,-5.6,;.64,-6.37,;-.7,-5.6,;-2.03,-6.37,;-3.36,-5.6,;-2.03,-7.91,;-.7,-4.06,;-2.03,-3.29,;-2.03,-1.75,;-.7,-.98,;-.7,.56,;-2.03,1.33,;-2.03,2.87,;-.7,3.64,;-.7,5.18,;-2.03,5.95,;-3.36,5.18,;-3.36,3.64,;-2.03,7.49,;-3.36,8.26,;-3.36,9.8,;-2.03,10.57,;-2.03,12.11,;-.7,9.8,;-.7,8.26,;-3.36,.56,;-4.7,1.33,;-3.36,-.98,;.64,-3.29,;1.97,-4.06,;3.3,-3.29,;4.64,-4.06,;4.64,-5.6,;5.97,-6.37,;7.3,-5.6,;8.12,-6.91,;8.84,-5.65,;7.3,-4.06,;5.97,-3.29,)| | ||
| Structure | 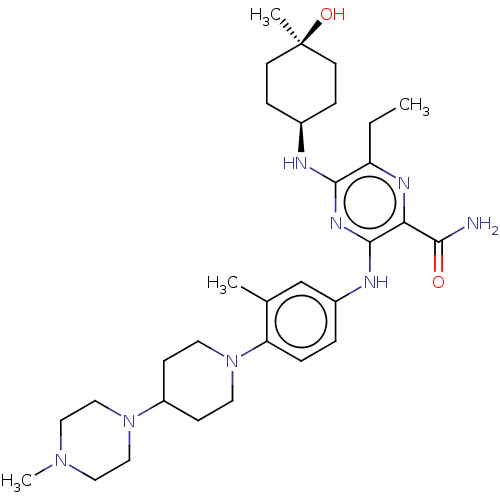
| ||