| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Protein mono-ADP-ribosyltransferase PARP15 [460-656,Y598L] |
|---|
| Ligand | BDBM199181 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Enzymatic Activity Assay |
|---|
| IC50 | 295±0.0 nM |
|---|
| Citation |  Venkannagari, H; Verheugd, P; Koivunen, J; Haikarainen, T; Obaji, E; Ashok, Y; Narwal, M; Pihlajaniemi, T; Lüscher, B; Lehtiö, L Small-Molecule Chemical Probe Rescues Cells from Mono-ADP-Ribosyltransferase ARTD10/PARP10-Induced Apoptosis and Sensitizes Cancer Cells to DNA Damage. Cell Chem Biol23:1251-1260 (2016) [PubMed] Article Venkannagari, H; Verheugd, P; Koivunen, J; Haikarainen, T; Obaji, E; Ashok, Y; Narwal, M; Pihlajaniemi, T; Lüscher, B; Lehtiö, L Small-Molecule Chemical Probe Rescues Cells from Mono-ADP-Ribosyltransferase ARTD10/PARP10-Induced Apoptosis and Sensitizes Cancer Cells to DNA Damage. Cell Chem Biol23:1251-1260 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Protein mono-ADP-ribosyltransferase PARP15 [460-656,Y598L] |
|---|
| Name: | Protein mono-ADP-ribosyltransferase PARP15 [460-656,Y598L] |
|---|
| Synonyms: | BAL3 | Human diphtheria toxin-like ADP-ribosyltransferase (Y598L) (ARTD7 or PARP15 (Y598L)) | PAR15_HUMAN | PARP15 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 22452.58 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q460N3[460-656,Y598L] |
|---|
| Residue: | 197 |
|---|
| Sequence: | MKKRDLSASLNFQSTFSMTTCNLPEHWTDMNHQLFCMVQLEPGQSEYNTIKDKFTRTCSS
YAIEKIERIQNAFLWQSYQVKKRQMDIKNDHKNNERLLFHGTDADSVPYVNQHGFNRSCA
GKNAVSYGKGTYFAVDASLSAKDTYSKPDSNGRKHMYVVRVLTGVFTKGRAGLVTPPPKN
PHNPTDLFDSVTNNTRS
|
|
|
|---|
| BDBM199181 |
|---|
| n/a |
|---|
| Name | BDBM199181 |
|---|
| Synonyms: | 4-[(4-Carbamoylcyclohexyl)oxy]cyclohexane-1-carboxamide (OUL35) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H12N2O3 |
|---|
| Mol. Mass. | 256.2567 |
|---|
| SMILES | NC(=O)c1ccc(Oc2ccc(cc2)C(N)=O)cc1 |
|---|
| Structure | 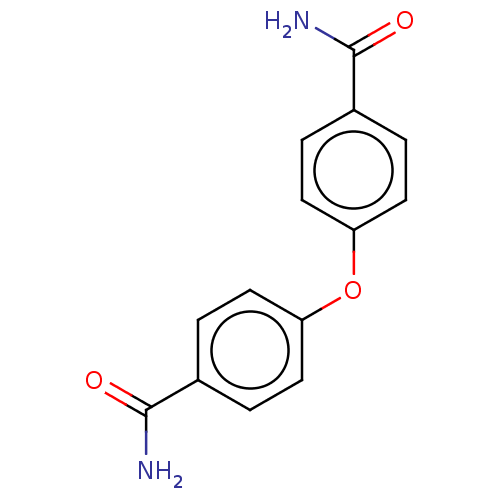
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Venkannagari, H; Verheugd, P; Koivunen, J; Haikarainen, T; Obaji, E; Ashok, Y; Narwal, M; Pihlajaniemi, T; Lüscher, B; Lehtiö, L Small-Molecule Chemical Probe Rescues Cells from Mono-ADP-Ribosyltransferase ARTD10/PARP10-Induced Apoptosis and Sensitizes Cancer Cells to DNA Damage. Cell Chem Biol23:1251-1260 (2016) [PubMed] Article
Venkannagari, H; Verheugd, P; Koivunen, J; Haikarainen, T; Obaji, E; Ashok, Y; Narwal, M; Pihlajaniemi, T; Lüscher, B; Lehtiö, L Small-Molecule Chemical Probe Rescues Cells from Mono-ADP-Ribosyltransferase ARTD10/PARP10-Induced Apoptosis and Sensitizes Cancer Cells to DNA Damage. Cell Chem Biol23:1251-1260 (2016) [PubMed] Article