| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Alpha-enolase |
|---|
| Ligand | BDBM204923 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Enolase Enzymatic Assays |
|---|
| pH | 7.4±n/a |
|---|
| IC50 | 53.2±0.0 nM |
|---|
| Comments | extracted |
|---|
| Citation |  Leonard, PG; Satani, N; Maxwell, D; Lin, YH; Hammoudi, N; Peng, Z; Pisaneschi, F; Link, TM; Lee, GR; Sun, D; Prasad, BA; Di Francesco, ME; Czako, B; Asara, JM; Wang, YA; Bornmann, W; DePinho, RA; Muller, FL SF2312 is a natural phosphonate inhibitor of enolase. Nat Chem Biol12:1053-1058 (2016) [PubMed] Article Leonard, PG; Satani, N; Maxwell, D; Lin, YH; Hammoudi, N; Peng, Z; Pisaneschi, F; Link, TM; Lee, GR; Sun, D; Prasad, BA; Di Francesco, ME; Czako, B; Asara, JM; Wang, YA; Bornmann, W; DePinho, RA; Muller, FL SF2312 is a natural phosphonate inhibitor of enolase. Nat Chem Biol12:1053-1058 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Alpha-enolase |
|---|
| Name: | Alpha-enolase |
|---|
| Synonyms: | 2-phospho-D-glycerate hydro-lyase | 4.2.1.11 | C-myc promoter-binding protein | ENO1 | ENO1L1 | ENOA_HUMAN | Enolase 1 | Enolase 1 (ENO1) | MBP-1 | MBPB1 | MPB-1 | MPB1 | NNE | Non-neural enolase | Phosphopyruvate hydratase | Plasminogen-binding protein |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 47171.43 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P06733 |
|---|
| Residue: | 434 |
|---|
| Sequence: | MSILKIHAREIFDSRGNPTVEVDLFTSKGLFRAAVPSGASTGIYEALELRDNDKTRYMGK
GVSKAVEHINKTIAPALVSKKLNVTEQEKIDKLMIEMDGTENKSKFGANAILGVSLAVCK
AGAVEKGVPLYRHIADLAGNSEVILPVPAFNVINGGSHAGNKLAMQEFMILPVGAANFRE
AMRIGAEVYHNLKNVIKEKYGKDATNVGDEGGFAPNILENKEGLELLKTAIGKAGYTDKV
VIGMDVAASEFFRSGKYDLDFKSPDDPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDD
WGAWQKFTASAGIQVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLA
QANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSK
AKFAGRNFRNPLAK
|
|
|
|---|
| BDBM204923 |
|---|
| n/a |
|---|
| Name | BDBM204923 |
|---|
| Synonyms: | phosphonoacetohydroxamate (PhAH) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C2H6NO5P |
|---|
| Mol. Mass. | 155.0465 |
|---|
| SMILES | ONC(=O)CP(O)(O)=O |
|---|
| Structure | 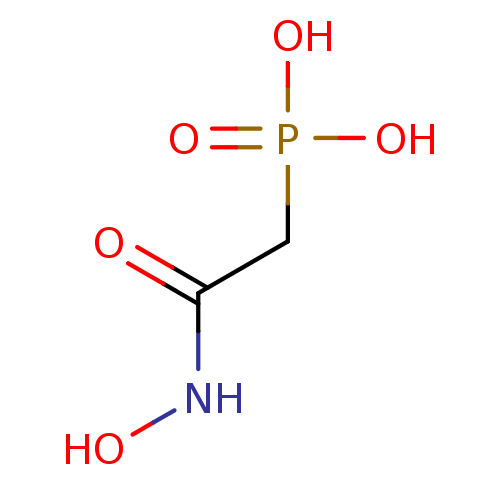
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Leonard, PG; Satani, N; Maxwell, D; Lin, YH; Hammoudi, N; Peng, Z; Pisaneschi, F; Link, TM; Lee, GR; Sun, D; Prasad, BA; Di Francesco, ME; Czako, B; Asara, JM; Wang, YA; Bornmann, W; DePinho, RA; Muller, FL SF2312 is a natural phosphonate inhibitor of enolase. Nat Chem Biol12:1053-1058 (2016) [PubMed] Article
Leonard, PG; Satani, N; Maxwell, D; Lin, YH; Hammoudi, N; Peng, Z; Pisaneschi, F; Link, TM; Lee, GR; Sun, D; Prasad, BA; Di Francesco, ME; Czako, B; Asara, JM; Wang, YA; Bornmann, W; DePinho, RA; Muller, FL SF2312 is a natural phosphonate inhibitor of enolase. Nat Chem Biol12:1053-1058 (2016) [PubMed] Article