

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Isoform C of Bromodomain-containing protein 4 (Short) | ||
| Ligand | BDBM50380682 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | AlphaScreen BRD Binding Assay | ||
| pH | 7.5±n/a | ||
| Temperature | 298.15±n/a K | ||
| IC50 | 29.4±0.0 nM | ||
| Comments | extracted | ||
| Citation |  Tanaka, M; Roberts, JM; Seo, HS; Souza, A; Paulk, J; Scott, TG; DeAngelo, SL; Dhe-Paganon, S; Bradner, JE Design and characterization of bivalent BET inhibitors. Nat Chem Biol12:1089-1096 (2016) [PubMed] Article Tanaka, M; Roberts, JM; Seo, HS; Souza, A; Paulk, J; Scott, TG; DeAngelo, SL; Dhe-Paganon, S; Bradner, JE Design and characterization of bivalent BET inhibitors. Nat Chem Biol12:1089-1096 (2016) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Isoform C of Bromodomain-containing protein 4 (Short) | |||
| Name: | Isoform C of Bromodomain-containing protein 4 (Short) | ||
| Synonyms: | BET bromodomain 4 (BRD4) | BRD4 | BRD4_HUMAN | Bromodomain-containing protein 4 (Short) | HUNK1 | ||
| Type: | n/a | ||
| Mol. Mass.: | 80477.46 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O60885-2 | ||
| Residue: | 722 | ||
| Sequence: |
| ||
| BDBM50380682 | |||
| n/a | |||
| Name | BDBM50380682 | ||
| Synonyms: | CHEMBL2017291 | I-BET151 (16) | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H21N5O3 | ||
| Mol. Mass. | 415.4445 | ||
| SMILES | COc1cc2c3n([C@H](C)c4ccccn4)c(=O)[nH]c3cnc2cc1-c1c(C)noc1C |r,wD:7.7,(46.55,-39.62,;46.55,-41.16,;47.89,-41.93,;49.21,-41.16,;50.55,-41.92,;51.88,-41.15,;52.18,-39.65,;51.41,-38.32,;49.87,-38.32,;52.18,-36.98,;51.41,-35.66,;52.17,-34.32,;53.72,-34.32,;54.49,-35.66,;53.72,-36.99,;53.7,-39.47,;54.46,-38.13,;54.34,-40.87,;53.21,-41.9,;53.22,-43.46,;51.89,-44.23,;50.55,-43.47,;49.22,-44.25,;47.88,-43.47,;46.55,-44.24,;46.51,-45.79,;47.74,-46.72,;45.04,-46.23,;44.16,-44.97,;45.09,-43.74,;44.64,-42.27,)| | ||
| Structure | 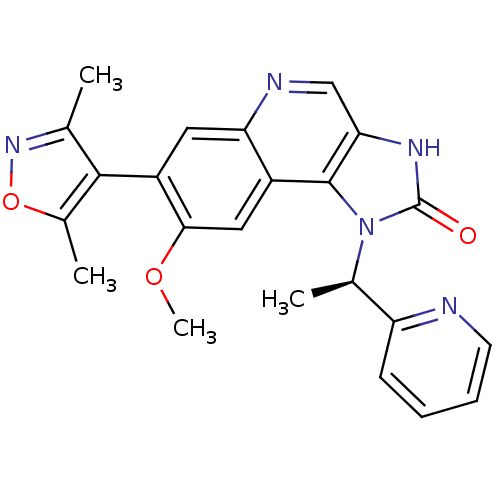
| ||