| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-dependent Clp protease ATP-binding subunit ClpC1 [1-145,F80Y] |
|---|
| Ligand | BDBM213240 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Isothermal Titration Calorimetry (ITC) |
|---|
| pH | 7.5±0 |
|---|
| Temperature | 298.15±0 K |
|---|
| Kd | 124±12 nM |
|---|
| Citation |  Vasudevan, D; Rao, SP; Noble, CG Structural basis of mycobacterial inhibition by cyclomarin A J Biol Chem288:30883-91 (2013) [PubMed] Article Vasudevan, D; Rao, SP; Noble, CG Structural basis of mycobacterial inhibition by cyclomarin A J Biol Chem288:30883-91 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| ATP-dependent Clp protease ATP-binding subunit ClpC1 [1-145,F80Y] |
|---|
| Name: | ATP-dependent Clp protease ATP-binding subunit ClpC1 [1-145,F80Y] |
|---|
| Synonyms: | CLPC1_MYCTU | Caseinolytic protein C1 (ClpC1 NTD F80Y) | clpC1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 15998.24 |
|---|
| Organism: | Mycobacterium tuberculosis |
|---|
| Description: | M. tuberculosis ClpC1 N-terminal domain mutant F80Y |
|---|
| Residue: | 145 |
|---|
| Sequence: | MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLESLGISLEGVR
SQVEEIIGQGQQAPSGHIPYTPRAKKVLELSLREALQLGHNYIGTEHILLGLIREGEGVA
AQVLVKLGAELTRVRQQVIQLLSGY
|
|
|
|---|
| BDBM213240 |
|---|
| n/a |
|---|
| Name | BDBM213240 |
|---|
| Synonyms: | Cyclomarin A1 (CymA1) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C59H92N10O11 |
|---|
| Mol. Mass. | 1117.4222 |
|---|
| SMILES | [#6]-[#8]-[#6@@H](-[#6@@H]-1-[#7]-[#6](=O)-[#6@H](-[#6])-[#7]-[#6](=O)-[#6@H](-[#6]-[#6@@H](-[#6])-[#6]-[#8])-[#7](-[#6])-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@@H](-[#7]-[#6](=O)-[#6@H](-[#6]-[#6](-[#6])-[#6])-[#7](-[#6])-[#6](=O)-[#6@@H](-[#7]-[#6]-1=O)-[#6](-[#6])-[#6])-[#6@H](-[#6])\[#6]=[#6](/[#6])-[#6])-[#6@H](-[#8])-c1cn(c2ccccc12)C([#6])([#6])[#6@@H](-[#8])-[#6]-[#7]-[#6]-[#6]-[#6]-[#7])-c1ccccc1 |r| |
|---|
| Structure | 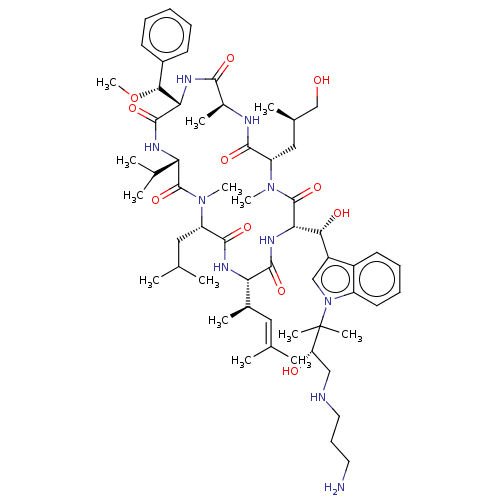
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Vasudevan, D; Rao, SP; Noble, CG Structural basis of mycobacterial inhibition by cyclomarin A J Biol Chem288:30883-91 (2013) [PubMed] Article
Vasudevan, D; Rao, SP; Noble, CG Structural basis of mycobacterial inhibition by cyclomarin A J Biol Chem288:30883-91 (2013) [PubMed] Article