| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histidine-binding periplasmic protein [1-238] |
|---|
| Ligand | BDBM7953 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Isothermal Titration Calorimetry (ITC) |
|---|
| pH | 7±0 |
|---|
| Temperature | 303.15±0 K |
|---|
| Kd | 114±16 nM |
|---|
| Citation |  Chu, BC; DeWolf, T; Vogel, HJ Role of the two structural domains from the periplasmic Escherichia coli histidine-binding protein HisJ. J Biol Chem288:31409-22 (2013) [PubMed] Article Chu, BC; DeWolf, T; Vogel, HJ Role of the two structural domains from the periplasmic Escherichia coli histidine-binding protein HisJ. J Biol Chem288:31409-22 (2013) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histidine-binding periplasmic protein [1-238] |
|---|
| Name: | Histidine-binding periplasmic protein [1-238] |
|---|
| Synonyms: | HISJ_ECOLI | Histidine-binding periplasmic protein (HisJ) | hisJ |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 25923.66 |
|---|
| Organism: | Escherichia coli (Enterobacteria) |
|---|
| Description: | E. coli HisJ truncation (1-238 aa) without periplasmic signal sequence |
|---|
| Residue: | 238 |
|---|
| Sequence: | MKKLVLSLSLVLAFSSATAAFAAIPQNIRIGTDPTYAPFESKNSQGELVGFDIDLAKELC
KRINTQCTFVENPLDALIPSLKAKKIDAIMSSLSITEKRQQEIAFTDKLYAADSRLVVAK
NSDIQPTVESLKGKRVGVLQGTTQETFGNEHWAPKGIEIVSYQGQDNIYSDLTAGRIDAA
FQDEVAASEGFLKQPVGKDYKFGGPSVKDEKLFGVGTGMGLRKEDNELREALNKAFAE
|
|
|
|---|
| BDBM7953 |
|---|
| n/a |
|---|
| Name | BDBM7953 |
|---|
| Synonyms: | (2S)-2-amino-3-(1H-imidazol-4-yl)propanoic acid | Histidine | Imidazole C-4(5) deriv. 5 | L-2-Amino-3-(4-imidazolyl)propionic acid | L-Histidine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C6H9N3O2 |
|---|
| Mol. Mass. | 155.1546 |
|---|
| SMILES | N[C@@H](Cc1cnc[nH]1)C(O)=O |r| |
|---|
| Structure | 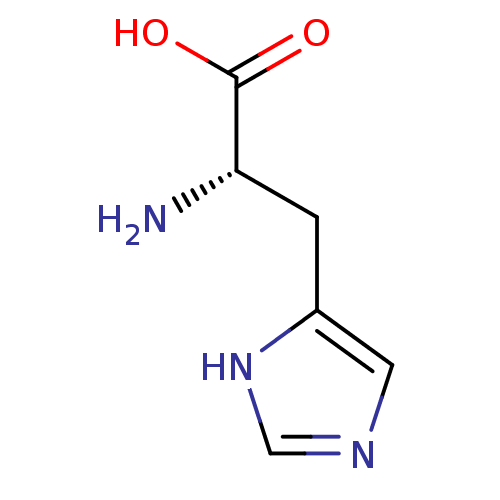
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chu, BC; DeWolf, T; Vogel, HJ Role of the two structural domains from the periplasmic Escherichia coli histidine-binding protein HisJ. J Biol Chem288:31409-22 (2013) [PubMed] Article
Chu, BC; DeWolf, T; Vogel, HJ Role of the two structural domains from the periplasmic Escherichia coli histidine-binding protein HisJ. J Biol Chem288:31409-22 (2013) [PubMed] Article