| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 6-phosphogluconate dehydrogenase, decarboxylating |
|---|
| Ligand | BDBM48320 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | In Vitro Assay |
|---|
| Ki | 2111300±697900 nM |
|---|
| IC50 | 3270000±0.0 nM |
|---|
| Citation |  Akkemik, E; Budak, H; Ciftci, M Effects of some drugs on human erythrocyte 6-phosphogluconate dehydrogenase: an in vitro study. J Enzyme Inhib Med Chem25:476-9 (2010) [PubMed] Article Akkemik, E; Budak, H; Ciftci, M Effects of some drugs on human erythrocyte 6-phosphogluconate dehydrogenase: an in vitro study. J Enzyme Inhib Med Chem25:476-9 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 6-phosphogluconate dehydrogenase, decarboxylating |
|---|
| Name: | 6-phosphogluconate dehydrogenase, decarboxylating |
|---|
| Synonyms: | 6-phosphogluconate dehydrogenase | 6-phosphogluconate dehydrogenase (6PGD) | 6PGD_HUMAN | PGD | PGDH |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 53143.42 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P52209 |
|---|
| Residue: | 483 |
|---|
| Sequence: | MAQADIALIGLAVMGQNLILNMNDHGFVVCAFNRTVSKVDDFLANEAKGTKVVGAQSLKE
MVSKLKKPRRIILLVKAGQAVDDFIEKLVPLLDTGDIIIDGGNSEYRDTTRRCRDLKAKG
ILFVGSGVSGGEEGARYGPSLMPGGNKEAWPHIKTIFQGIAAKVGTGEPCCDWVGDEGAG
HFVKMVHNGIEYGDMQLICEAYHLMKDVLGMAQDEMAQAFEDWNKTELDSFLIEITANIL
KFQDTDGKHLLPKIRDSAGQKGTGKWTAISALEYGVPVTLIGEAVFARCLSSLKDERIQA
SKKLKGPQKFQFDGDKKSFLEDIRKALYASKIISYAQGFMLLRQAATEFGWTLNYGGIAL
MWRGGCIIRSVFLGKIKDAFDRNPELQNLLLDDFFKSAVENCQDSWRRAVSTGVQAGIPM
PCFTTALSFYDGYRHEMLPASLIQAQRDYFGAHTYELLAKPGQFIHTNWTGHGGTVSSSS
YNA
|
|
|
|---|
| BDBM48320 |
|---|
| n/a |
|---|
| Name | BDBM48320 |
|---|
| Synonyms: | 4-amino-5-chloro-N-[2-(diethylamino)ethyl]-2-methoxy-benzamide;hydrochloride | 4-amino-5-chloro-N-[2-(diethylamino)ethyl]-2-methoxybenzamide;hydrochloride | 4-azanyl-5-chloranyl-N-[2-(diethylamino)ethyl]-2-methoxy-benzamide;hydrochloride | METOCLOPRAMIDE | METOCLOPRAMIDE HYDROCHLORIDE | MLS000069667 | Metochloropramide | SMR000058471 | US11147820, Compound Metoclopramide | US11717500, Compound Metoclopramide | US9132134, Metoclopramide | cid_23659 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H22ClN3O2 |
|---|
| Mol. Mass. | 299.796 |
|---|
| SMILES | CCN(CC)CCNC(=O)c1cc(Cl)c(N)cc1OC |
|---|
| Structure | 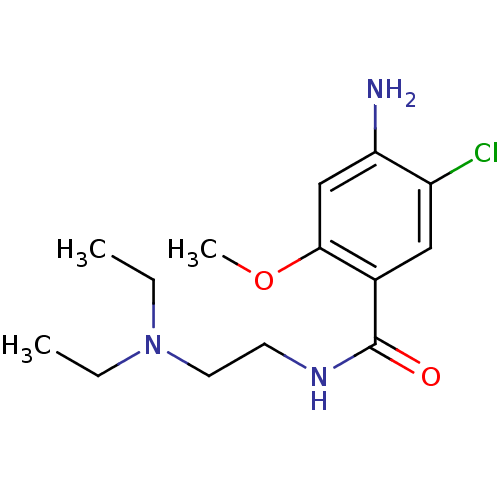
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Akkemik, E; Budak, H; Ciftci, M Effects of some drugs on human erythrocyte 6-phosphogluconate dehydrogenase: an in vitro study. J Enzyme Inhib Med Chem25:476-9 (2010) [PubMed] Article
Akkemik, E; Budak, H; Ciftci, M Effects of some drugs on human erythrocyte 6-phosphogluconate dehydrogenase: an in vitro study. J Enzyme Inhib Med Chem25:476-9 (2010) [PubMed] Article