null
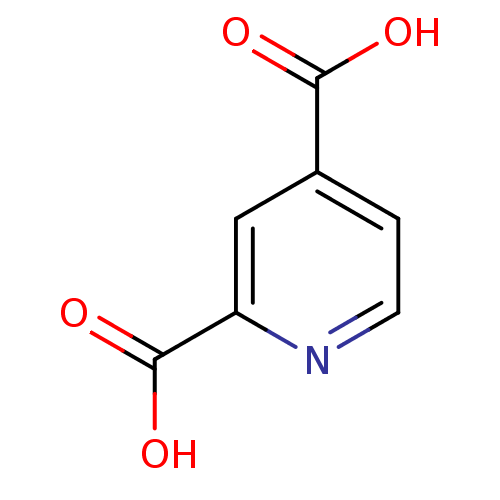
SMILES OC(=O)c1ccnc(c1)C(O)=O
InChI Key InChIKey=MJIVRKPEXXHNJT-UHFFFAOYSA-N
PDB links: 16 PDB IDs match this monomer. 18 PDB IDs contain this monomer as substructures. 18 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 26113
Found 3 hits for monomerid = 26113
TargetProlyl 4-hydroxylase(Paramecium bursaria Chlorella virus 1)
University of Oxford
Curated by ChEMBL
University of Oxford
Curated by ChEMBL
Affinity DataKi: 3.28E+3nMAssay Description:Inhibition of N-terminal His6-tagged recombinant Paramecium bursaria chlorella virus 1 CPH expressed in Escherichia coli Rosetta 2 (DE3) cells pre-in...More data for this Ligand-Target Pair
TargetProlyl 4-hydroxylase(Paramecium bursaria Chlorella virus 1)
University of Oxford
Curated by ChEMBL
University of Oxford
Curated by ChEMBL
Affinity DataIC50: 5.30E+3nMAssay Description:Inhibition of N-terminal His6-tagged recombinant Paramecium bursaria chlorella virus 1 CPH expressed in Escherichia coli Rosetta 2 (DE3) cells pre-in...More data for this Ligand-Target Pair
Affinity DataIC50: 7.00E+3nMAssay Description:Inhibition of recombinant human PHD2 using 2OG as substrate and Fe2 as co-factor assessed as hydroxylation incubated for 15 mins in presence of L-asc...More data for this Ligand-Target Pair