null
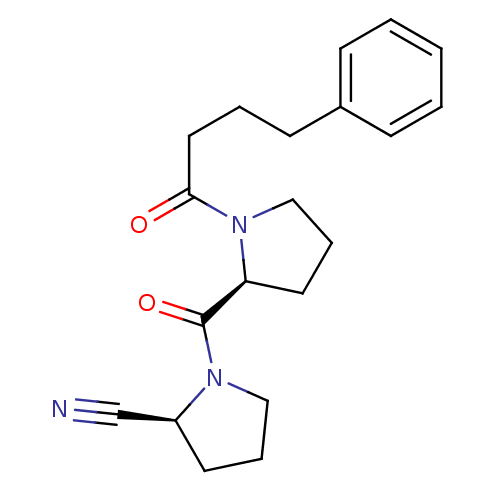
SMILES O=C(CCCc1ccccc1)N1CCC[C@H]1C(=O)N1CCC[C@H]1C#N
InChI Key InChIKey=SPXFAUXQZWJGCJ-ROUUACIJSA-N
PDB links: 1 PDB ID matches this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 50155838
Found 4 hits for monomerid = 50155838
Affinity DataKi: 0.0230nMAssay Description:Inhibition of prolyl oligopeptidase (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 0.100nMAssay Description:Inhibition of porcine prolyl oligopeptidase using Z-Gly-Pro-AMC as substrate after 60 mins by double-reciprocal plot analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 0.860nMAssay Description:Inhibition of recombinant porcine brain PREP expressed in Escherichia coli assessed as formation of 7-amido-4-methylcoumarin using Suc-Gly-Pro-amido-...More data for this Ligand-Target Pair
Affinity DataIC50: 0.300nMAssay Description:Inhibition of PREP in C57BL/6 mouse cortex homogenate assessed as formation of 7-amido-4-methylcoumarin using Suc-Gly-Pro-amido-4-methylcoumarin as s...More data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)