null
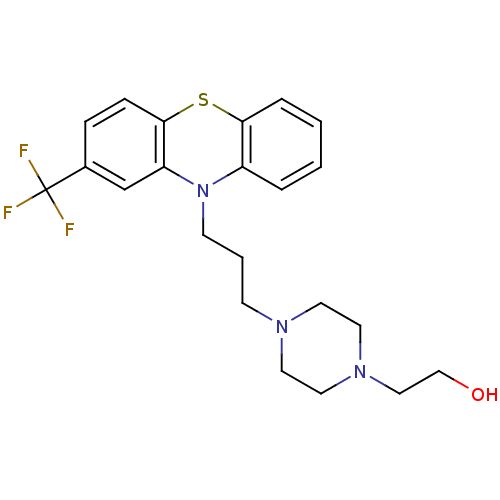
SMILES OCCN1CCN(CCCN2c3ccccc3Sc3ccc(cc23)C(F)(F)F)CC1
InChI Key InChIKey=PLDUPXSUYLZYBN-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 78433
Found 3 hits for monomerid = 78433
TargetATP-dependent translocase ABCB1(Homo sapiens (Human))
Schering-Plough Research Institute
Curated by ChEMBL
Schering-Plough Research Institute
Curated by ChEMBL
Affinity DataIC50: 1.04E+4nMAssay Description:TP_TRANSPORTER: inhibition of Rhodamine 123 efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
TargetATP-dependent translocase ABCB1(Homo sapiens (Human))
Schering-Plough Research Institute
Curated by ChEMBL
Schering-Plough Research Institute
Curated by ChEMBL
Affinity DataIC50: 6.50E+3nMAssay Description:TP_TRANSPORTER: inhibition of Daunorubicin efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair
TargetATP-dependent translocase ABCB1(Homo sapiens (Human))
Schering-Plough Research Institute
Curated by ChEMBL
Schering-Plough Research Institute
Curated by ChEMBL
Affinity DataIC50: 5.70E+3nMAssay Description:TP_TRANSPORTER: inhibition of LDS-751 efflux in NIH-3T3-G185 cellsMore data for this Ligand-Target Pair