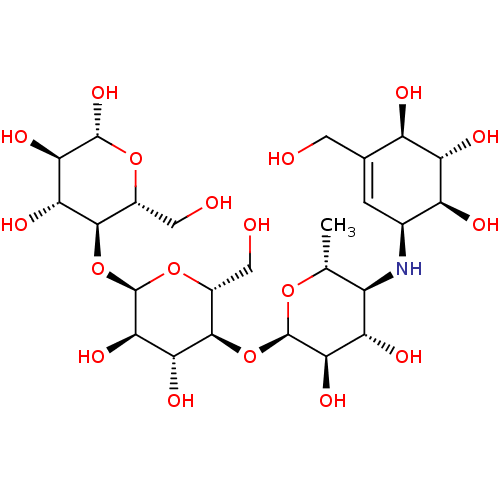
null
SMILES C[C@H]1O[C@H](O[C@@H]2[C@@H](CO)O[C@H](O[C@@H]3[C@@H](CO)O[C@@H](O)[C@H](O)[C@H]3O)[C@H](O)[C@H]2O)[C@H](O)[C@@H](O)[C@@H]1N[C@H]1C=C(CO)[C@@H](O)[C@H](O)[C@H]1O
InChI Key InChIKey=XUFXOAAUWZOOIT-SXARVLRPSA-N
PDB links: 2 PDB IDs match this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50333465
Found 2 hits for monomerid = 50333465
TargetSucrase-isomaltase, intestinal(Rattus norvegicus (Rat))
Chulalongkorn University
Curated by ChEMBL
Chulalongkorn University
Curated by ChEMBL
Affinity DataIC50: 2.30E+3nMAssay Description:Inhibition of rat intestine sucrase using sucrose as substrate incubated for 10 mins prior to substrate addition measured after 40 mins by glucose ox...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMAssay Description:Inhibition of rat intestine maltase using maltose as substrate incubated for 10 mins prior to substrate addition measured after 40 mins by glucose ox...More data for this Ligand-Target Pair