null
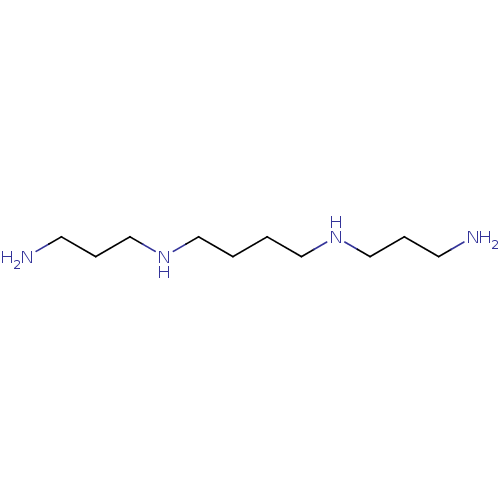
SMILES NCCCNCCCCNCCCN
InChI Key InChIKey=PFNFFQXMRSDOHW-UHFFFAOYSA-N
PDB links: 216 PDB IDs match this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 79403
Found 3 hits for monomerid = 79403
TargetGlutamate receptor ionotropic, NMDA 2B(Homo sapiens (Human))
University of Bristol
Curated by ChEMBL
University of Bristol
Curated by ChEMBL
Affinity DataEC50: 1.25E+5nMAssay Description:Positive allosteric modulation of GluN2B receptor (unknown origin) in hippocampal neurons assessed as increase in glycine-induced channel current by ...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2B(Rattus norvegicus (Rat))
University of Bristol
Curated by ChEMBL
University of Bristol
Curated by ChEMBL
Affinity DataEC50: 8.10E+4nMAssay Description:Positive allosteric modulation of GluN2B receptor in rat spinal cord neurons assessed as increase in glycine-induced channel current by two electrode...More data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, NMDA 2B(Homo sapiens (Human))
University of Bristol
Curated by ChEMBL
University of Bristol
Curated by ChEMBL
Affinity DataEC50: 1.27E+5nMAssay Description:Positive allosteric modulation of GluN2B receptor (unknown origin) expressed in xenopus laevis oocytes assessed as increase in glycine-induced channe...More data for this Ligand-Target Pair