null
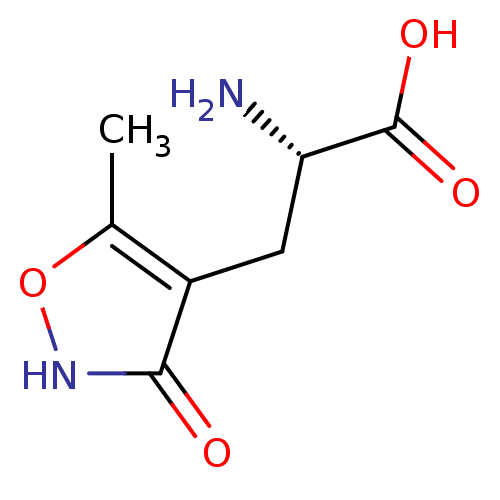
SMILES Cc1o[nH]c(=O)c1C[C@H](N)C(O)=O
InChI Key InChIKey=UUDAMDVQRQNNHZ-YFKPBYRVSA-N
PDB links: 8 PDB IDs match this monomer. 4 PDB IDs contain this monomer as substructures. 4 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 17662
Found 2 hits for monomerid = 17662
Affinity DataKi: 1.15E+3nMAssay Description:Displacement of [3H[SYM2081 from rat recombinant GluK1(Q)1b expressed in sS9 cellsMore data for this Ligand-Target Pair
TargetGlutamate receptor ionotropic, kainate 1(Homo sapiens (Human))
The Danish University of Pharmaceutical Sciences
Curated by ChEMBL
The Danish University of Pharmaceutical Sciences
Curated by ChEMBL
Affinity DataKi: 1.15E+3nMAssay Description:Binding affinity for kainate Glutamate receptor GluR5 expressed in Sf9 cellsMore data for this Ligand-Target Pair