null
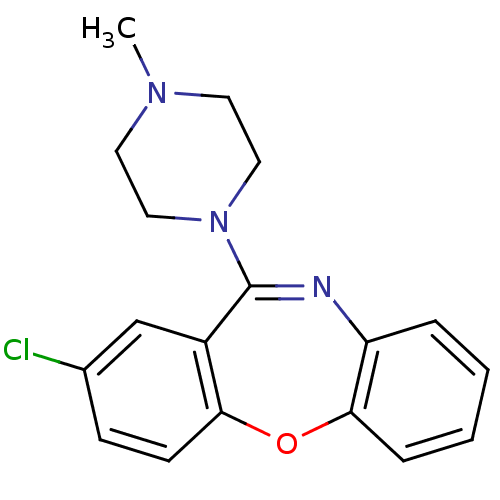
SMILES CN1CCN(CC1)C1=Nc2ccccc2Oc2ccc(Cl)cc12
InChI Key InChIKey=XJGVXQDUIWGIRW-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 22871
Found 4 hits for monomerid = 22871
TargetHistamine H1 receptor(Homo sapiens (Human))
Mayo Clinic and Foundation
Curated by PDSP Ki Database
Mayo Clinic and Foundation
Curated by PDSP Ki Database
TargetHistamine H1 receptor(Homo sapiens (Human))
Mayo Clinic and Foundation
Curated by PDSP Ki Database
Mayo Clinic and Foundation
Curated by PDSP Ki Database
TargetHistamine H1 receptor(Homo sapiens (Human))
Mayo Clinic and Foundation
Curated by PDSP Ki Database
Mayo Clinic and Foundation
Curated by PDSP Ki Database
Affinity DataIC50: 1.00E+3nMAssay Description:Binding affinity towards histamine H1 receptor from rat brain membranes using [3H]-mepyramine as radioligandMore data for this Ligand-Target Pair