null
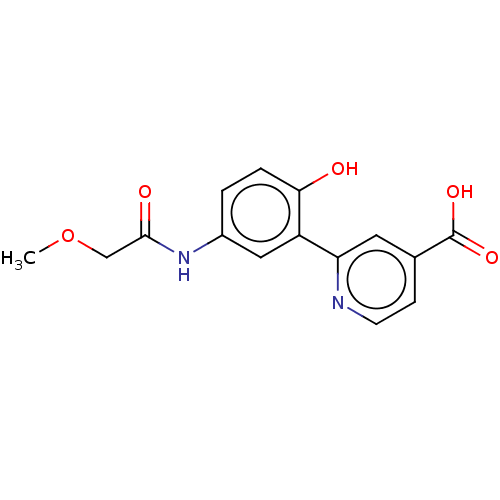
SMILES COCC(=O)Nc1ccc(O)c(c1)-c1cc(ccn1)C(O)=O
InChI Key InChIKey=HHHINYOXTBYXCI-UHFFFAOYSA-N
PDB links: 1 PDB ID matches this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 8 hits for monomerid = 50151399
Found 8 hits for monomerid = 50151399
Affinity DataKi: 683nMAssay Description:Competitive inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using A...More data for this Ligand-Target Pair
Affinity DataIC50: 1.42E+4nMAssay Description:Competitive inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using 5...More data for this Ligand-Target Pair
Affinity DataIC50: 1.49E+4nMAssay Description:Competitive inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using 2...More data for this Ligand-Target Pair
Affinity DataIC50: 3.30E+3nMAssay Description:Competitive inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using 5...More data for this Ligand-Target Pair
Affinity DataIC50: 4.70E+4nMAssay Description:Inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using ARTKQTARK(Me3...More data for this Ligand-Target Pair
Affinity DataIC50: 3.80E+3nMAssay Description:Inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using biotinylated ...More data for this Ligand-Target Pair
Affinity DataIC50: 140nMAssay Description:Inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using biotinylated ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.40E+3nMAssay Description:Inhibition of N-terminal His6-tagged KDM4C (1 to 352 residues) (unknown origin) expressed in Escherichia coli Rosetta 2(DE3)pLysS using biotinylated ...More data for this Ligand-Target Pair