null
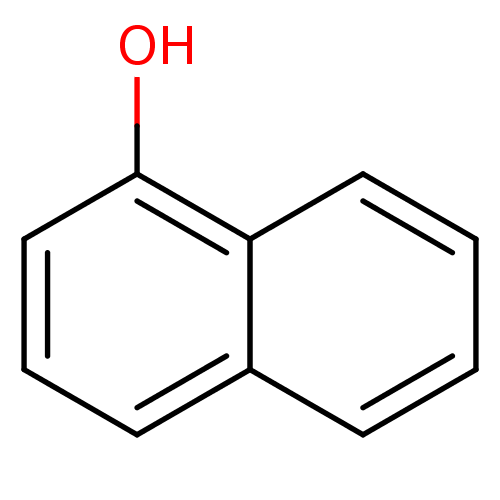
SMILES Oc1cccc2ccccc12
InChI Key InChIKey=KJCVRFUGPWSIIH-UHFFFAOYSA-N
PDB links: 7 PDB IDs match this monomer. 34 PDB IDs contain this monomer as substructures. 34 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 1 hit for monomerid = 23450
Found 1 hit for monomerid = 23450
Affinity DataKd: 4.30E+5nMAssay Description:The thermal-shift denaturation assay was performed on an iCycler iQ Real Time Detection System (BioRad) in 96-well iCycler iQ PCR plates sealed with ...More data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 2 hits for monomerid = 23450
Found 2 hits for monomerid = 23450
ITC DataΔG°: -5.30kcal/mole −TΔS°: 5.59kcal/mole ΔH°: -10.9kcal/mole logk: 6.73E+3
pH: 7.0 T: 30.00°C
pH: 7.0 T: 30.00°C
ITC DataΔG°: -7.63kcal/mole logk: 4.00E+5
pH: 7.0 T: 25.00°C
pH: 7.0 T: 25.00°C
 3D Structure (crystal)
3D Structure (crystal)