null
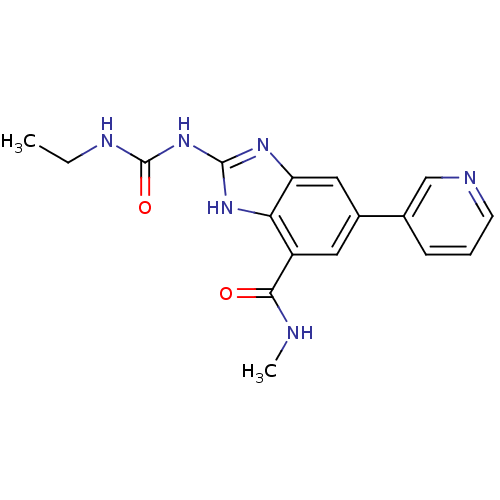
SMILES CCNC(=O)Nc1nc2cc(cc(C(=O)NC)c2[nH]1)-c1cccnc1
InChI Key InChIKey=VJBJRURYRWMPOS-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 24607
Found 3 hits for monomerid = 24607
Affinity DataKi: 5nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 26nM ΔG°: -10.5kcal/molepH: 7.5 T: 2°CAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair
Affinity DataKi: 150nMAssay Description:Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ...More data for this Ligand-Target Pair