null
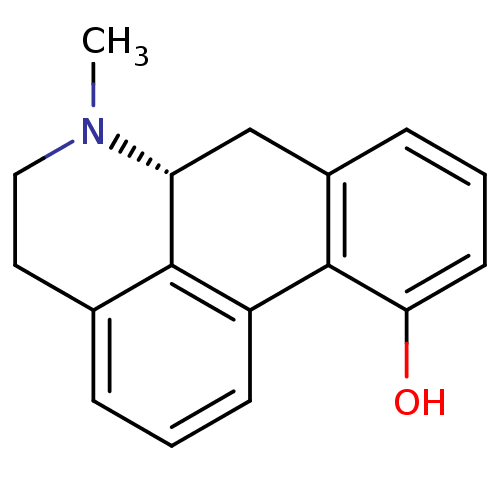
SMILES CN1CCc2cccc-3c2[C@H]1Cc1cccc(O)c-31
InChI Key InChIKey=PCGXWSCASZVBJT-CQSZACIVSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 17 hits for monomerid = 50016013
Found 17 hits for monomerid = 50016013
Affinity DataKi: 9.60nMAssay Description:Ability to displace [3H]-8-OH-DPAT binding to rat hippocampal 5-hydroxytryptamine 1A receptorMore data for this Ligand-Target Pair
Affinity DataKi: 9.60nMAssay Description:In vitro displacement of [3H]-8-OH-DPAT binding to rat hippocampal 5-hydroxytryptamine 1A receptorMore data for this Ligand-Target Pair
Affinity DataKi: 9.60nMAssay Description:In vitro affinity at 5-hydroxytryptamine 1A receptor of rat hippocampus by [3H]-8-OH-DPAT displacement.More data for this Ligand-Target Pair
Affinity DataKi: 20.4nMAssay Description:In vitro affinity at Dopamine receptor D1 of rat striatum by [3H]-SCH-23,390 displacement.More data for this Ligand-Target Pair
Affinity DataKi: 20.4nMAssay Description:In vitro displacement of [3H]-SCH-23,390 binding to rat striatal Dopamine receptor D1More data for this Ligand-Target Pair
Affinity DataKi: 20.4nMAssay Description:Ability to displace [3H]-SCH-23,390 binding to rat striatal Dopamine receptor D1More data for this Ligand-Target Pair
Target5-hydroxytryptamine receptor 1A(Homo sapiens (Human))
Chinese Academy of Sciences
Curated by ChEMBL
Chinese Academy of Sciences
Curated by ChEMBL
Affinity DataKi: 49nMAssay Description:Displacement of [3H]8-OH-DPAT from 5HT1A receptor expressed in CHO cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 58.5nMAssay Description:Ability to displace [3H]-raclopride binding to cloned human Dopamine receptor D2AMore data for this Ligand-Target Pair
Affinity DataKi: 58.5nMAssay Description:In vitro affinity at human cloned Dopamine receptor D2A by [3H]-raclopride displacement.More data for this Ligand-Target Pair
Affinity DataKi: 58.5nMAssay Description:Displacement of [3H]-raclopride from human Dopamine receptor D2AMore data for this Ligand-Target Pair
Affinity DataKi: 61nMAssay Description:Displacement of [3H]SCH23390 from dopamine D1 receptor expressed in HEK293 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataKi: 208nMAssay Description:Displacement of [3H]spiperone from dopamine D2 receptor expressed in HEK293 cells by liquid scintillation countingMore data for this Ligand-Target Pair
Affinity DataIC50: 58nMAssay Description:In vitro binding affinity towards rat Dopamine receptor D2 by [3H]-spiperone displacement.More data for this Ligand-Target Pair
Affinity DataIC50: 107nMAssay Description:In vitro binding affinity towards rat Dopamine receptor D1 by [3H]-SCH-23,390 displacement.More data for this Ligand-Target Pair
Affinity DataIC50: 58nMAssay Description:Displacement of [3H]raclopride from rat dopamine D2 receptorMore data for this Ligand-Target Pair
Affinity DataIC50: 110nMAssay Description:Displacement of [3H]SCH-23390 from rat dopamine D1 receptorMore data for this Ligand-Target Pair
Affinity DataEC50: 5.20E+3nMAssay Description:Ability to inhibit 5-HT sensitive,forskolin-stimulated adenylyl cyclase(FSC) activity in rat hippocampal membranes mediated through 5-hydroxytryptami...More data for this Ligand-Target Pair