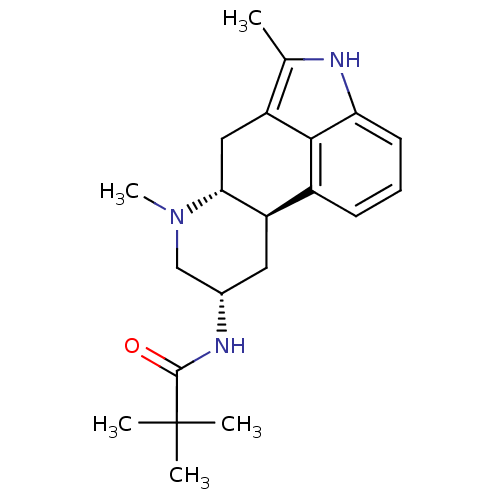
null
SMILES CN1C[C@H](C[C@H]2[C@H]1Cc1c(C)[nH]c3cccc2c13)NC(=O)C(C)(C)C
InChI Key InChIKey=ZNMWUMGOSWLWOH-FDQGKXFDSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2 hits for monomerid = 50061687
Found 2 hits for monomerid = 50061687
TargetD(2) dopamine receptor(Rattus norvegicus (rat))
Wyeth-Ayerst Research Laboratories
Curated by ChEMBL
Wyeth-Ayerst Research Laboratories
Curated by ChEMBL
Affinity DataKi: 0.900nMAssay Description:In vitro binding affinity to rat striatal Dopamine receptor D2 was determined using the agonist [3H]quinpirole to label the high affinity state (D2 h...More data for this Ligand-Target Pair
TargetD(2) dopamine receptor(Rattus norvegicus (rat))
Wyeth-Ayerst Research Laboratories
Curated by ChEMBL
Wyeth-Ayerst Research Laboratories
Curated by ChEMBL
Affinity DataKi: 1nMAssay Description:In vitro binding affinity to rat striatal Dopamine receptor D2 was determined using the antagonist [3H]spiperone + GTP to label the low affinity stat...More data for this Ligand-Target Pair